INTRODUCTION
Lindera angustifolia W. C. Cheng is a traditional medicinal plant and mainly distributed in the temperate zone of the China (Zhao et al., 2006). However, the species an extraordinary distribution, disjunctively isolated in western coastal islands of Korea, about 800 km northeast from China. In Korea, the plants are sometimes regarded as a variety, L. angustifolia var. glabra (Nakai) J. M. H. Shaw (Shaw, 2013). Therefore, the complete plastid genome sequence is an important basis for improving our understanding of the evolutionary process and solving taxonomic problems in inter/intra species within the genus Lindera.
MATERIALS AND METHODS
Plant material
A fresh leaf sample of L. angustifolia was collected from Daeyeonpyeongdo Island (37°39′44.0″N, 125°41′23.3″E), Yeonpyeong-ri, Ongjin-gun, Incheon. The voucher specimen of L. angustifolia was deposited at the Herbarium of the National Institute of Biological Resources in Korea (voucher number NIBRVP0000463020).
Chloroplast genome determination
The DNA library was constructed using Illumina Hi-seq platform (Macrogen, Seoul, Korea) and generated 56,426,712 raw reads (150 bp paired-end). The plastome was assembled using NOVOPlasty 4.3.1 (Dierckxsens et al., 2017), with the complete plastome of Chinese L. angustifolia (MG581438) (Jo et al., 2019) as the seed. The assembled plastome was checked using Geneious 11.0.9 (Kearse et al., 2012) by mapping 198,594 reads, resulting in a coverage of 150×. The annotation was conducted using Geneious 11.0.9 with the Chinese L. angustifolia (GenBank accession number MG581438) and L. glauca (GenBank accession number MG581443) (Jo et al., 2019) chloroplast complete genome as a reference. We manually corrected the start and stop codons as well as the intron/exon boundaries.
Phylogenetic analysis
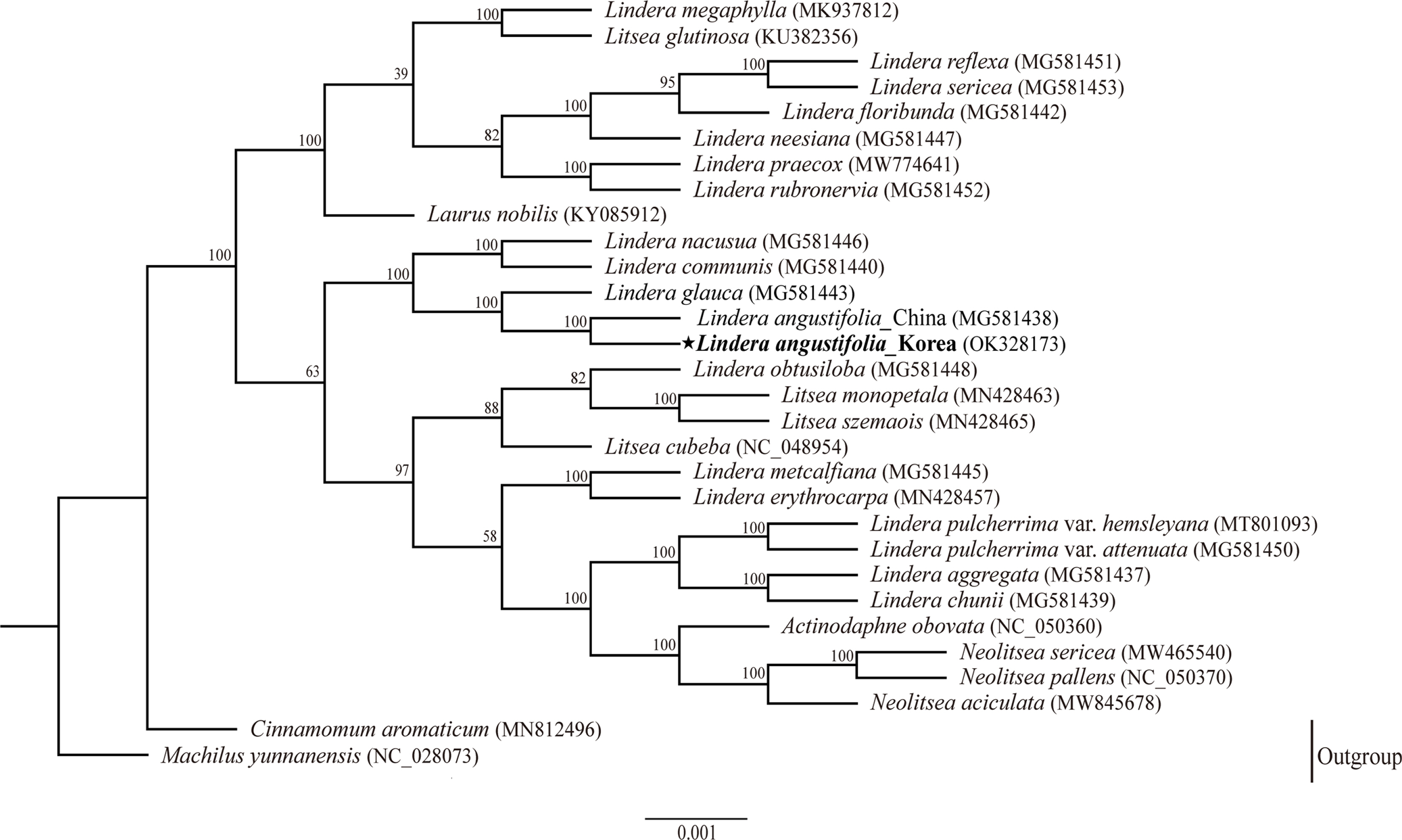
To reconstruct the phylogenetic tree, we downloaded 29 complete plastomes of Lauraceae (Lindera 18 taxa, Litsea 4 species, Neolitsea 3 species, and Laurus, Actinodaphne one species each, and two outgroups Cinnamomum and Machilus separately one species) from the NCBI database, including recently reported genome from Korea (Park and Cheon, 2021). The alignments were performed using MAFFT (Katoh and Toh, 2010). The maximum likelihood tree analysis was performed on a data set that includes 75 common genes extracted from 30 species with RAxML v.8.0 (Stamatakis, 2014) using default parameters and 1,000 bootstrap replicates. For the RAxML tree, the general time-reversible model of nucleotide substation was used with the Gamma model of rate heterogeneity.
RESULTS AND DISCUSSION
The novel annotated complete plastid genome of L. angustifolia is openly available in the NCBI GenBank at https://www.ncbi.nlm.nih.gov/ under the accession number OK328173. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA790663, SRR17287206, and SAMN24219956, respectively.
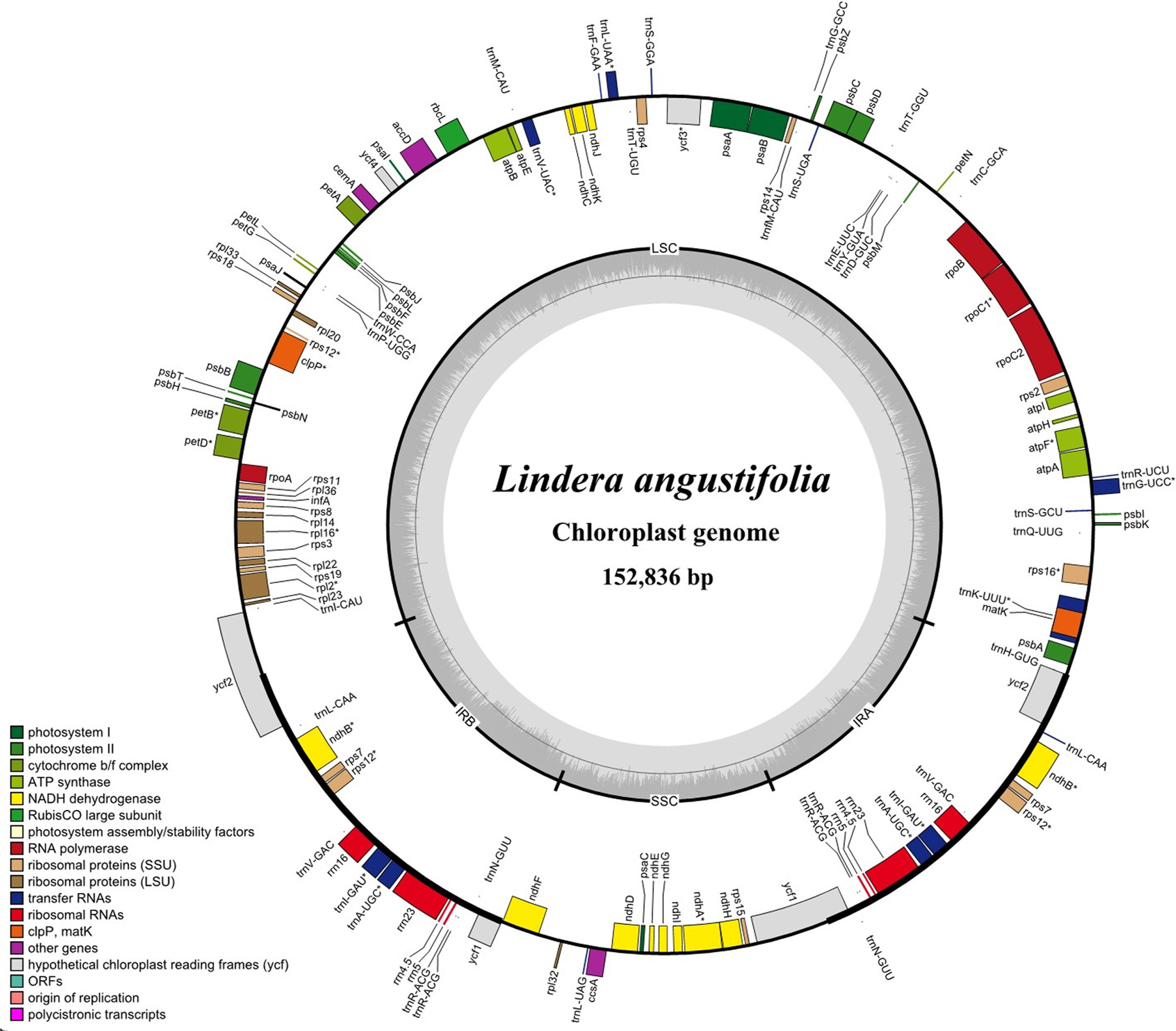
The complete chloroplast genome of L. angustifolia is 152,836 bp in length and has four subregions: 93,726 bp in the large single-copy (LSC) region and 18,946 bp in small single-copy (SSC) region are separated by two inverted repeat (IR) regions of 20,082 bp (Fig. 1). The overall GC content of L. angustifolia is 39.2% and those in the LSC, SSC, and IR regions are 38.0%, 33.9%, and 44.4%, respectively. A total 125 genes were detected including 81 protein-coding genes, 36 tRNA genes, and eight rRNA genes.
Among the genes, 18 genes (rps16, atpF, rpoC1, petB, petD, rpl16, rpl2, ndhB×2, ndhA, trnK-UUU, trnG-UCC, trnL-UAA, trnV-UAC, trnI-GAU×2, and trnA-UGC×2) contain a single intron and three genes (ycf3, clpP, and rps12) have two introns.
As a result of comparing the complete chloroplast genomes of Chinese and Korean L. angustifolia, the total genome size is 152,836 bp and 152,832 bp long respectively, with a difference of only 4 bp. In addition, the two genes (ycf1 and ycf2) in the IR regions and one gene (rpl22) in the LSC region are pseudogenized in both Chinese and Korean samples. The nucleotide sequences of the 1,372 bp of 3′– ycf1, 3,162 bp of 5′–ycf2, and 387 bp of 5′–rpl22 were truncated at the LSC and IR boundaries separately. Furthermore, while five RNA editing genes (psbL, rpl2, ndhB×2, and ndhD) were identified in L. angustifolia from China, the “ndhD” gene was not recognized as an RNA editing site in that Korean individual.
The phylogenetic tree (RAxML) constructed for 30 species of the Lauraceae family shows that Korean L. angustifolia is most closely related to Chinese L. angustifolia with strong (100%) bootstrap support and forms a sister group of L. glauca (Fig. 2). The additional plastome sequence will be a valuable, fundamental tool for future studies of phylogenetic relationships among species of Lindera, and will provide a useful resource for researching geographically genetic variations.