Abstract
Rhododendron caucasicum Pall. is a shrub distributed in the mountainous areas of the Caucasus from northeastern Türkiye towards the Caspian Sea. This study reports the first complete chloroplast genome sequence of R. caucasicum. The plastome is 199,487 base pairs (bp) long and exhibits a typical quadripartite structure comprising a large single-copy region of 107,645 bp, a small single-copy region of 2,598 bp, and a pair of identical inverted repeat regions of 44,622 bp each. It contains 143 genes, comprising 93 protein-coding genes, 42 tRNA genes, and eight rRNA genes. The large chloroplast genome size is likely due to the expansion of inverted repeats. A phylogenetic analysis of chloroplast genomes with other Rhododendron species supports previously recognized infrageneric relationship.
INTRODUCTION
Rhododendron is the largest woody plant genus in the Northern Hemisphere, comprising over 1,000 species (Frodin, 2004). A recent study indicated that the genus Rhododendron first originated in northeast Asia in the Paleocene and then dispersed to North America in the late Eocene and Oligocene (Shrestha et al., 2018). However, the contemporary species diversity of Rhododendron is mainly due to extensive speciation in the tropical and subtropical regions of southern China, south Asia, and the Malay Archipelago during the 30–10 MYA period (Milne et al., 2010; Shrestha et al., 2018). A recent molecular study divided the genus Rhododendron into five subgenera and 11 sections (Xia et al., 2022).
The chloroplast genome has been extensively used to clarify phylogenetic relationships from the species level to deeper levels (Gitzendanner et al., 2018; Li et al., 2019; Fan et al., 2021). Chloroplast genomes are the one of best molecular markers in plant phylogenetic studies due to their abundance and lack of recombination with appropriate mutation rates. Moreover, despite some exceptions, the maternal inheritance of chloroplasts contributes to its role as a key player in identifying ancient hybrid phenomena with comparison of the phylogenetic relationships of nuclear genes (Kawabe et al., 2018; Liu et al., 2022). Due to the high singlecell copy number and small genome size (120–160 kb) of plant chloroplasts, fast and cost-effective genome skimming is sufficient to obtain fully annotated whole genome sequences of the chloroplast.
Rhododendron caucasicum Pall. is a shrub distributed in the mountainous areas of the Caucasus from northeastern Türkiye towards the Caspian Sea. This species is phylogenetically closely related to R. aureum Georgi and R. brachycarpum D. Don ex G. Don, found in Northeast Asia (Milne, 2004). The disjunct distribution of R. caucasicum from R. aureum and R. brachycarpum and their phylogenetic closeness show that R. caucasicum is a rare case of a tertiary relict species in southwest Eurasia. Here, we report the complete chloroplast genome sequence of R. caucasicum. The chloroplast genome of R. caucasicum will aid further investigation into the biogeography of this species group.
MATERIALS AND METHODS
Rhododendron caucasicum was sampled at approximately 2,500 m in the timberline area of Tsratskharo Pass, close to Bakuriani, Samtskhe-Yavakheti, Georgia, by R. W. Bussmann in August 2022. The voucher specimen (RBU-19784) was deposited at the Herbarium of the National Institute of Biological Resources (KB) and the National Herbarium of Georgia (TBI). Genomic DNA was extracted from the dried leaves taken from the specimens using the cetyltrimethylammonium bromide method (Doyle and Doyle, 1987) and verified by 1% agarose gel electrophoresis. The DNA library was constructed using a TruSeq DNA Nano Kit for a 350-bp insert size according to the manufacturer’s instructions (Illumina Inc., San Diego, CA, USA). Whole-genome sequencing was performed using the Illumina NovaSeq6000 platform (DNA Link Inc., Seoul, Korea). We retrieved 7.3 Gb of raw reads (150 bp paired-end reads), which were quality-trimmed using the Trimmomatic tool (Bolger et al., 2014). De novo assembly was performed with Velvet v1.2.19 (Zerino and Birney, 2008), and the obtained contigs were used to construct a draft genome with the R. delavayi Franch. chloroplast genome (GenBank accession no. MN711645) as a reference. The genome sequence was confirmed by aligning the raw reads against the assembled genome using BWA v0.7.17 and SAMtools v1.9 (Li, 2013). The gaps were closed using GapCloser v1.12 (Zhao et al., 2011). Annotation of the chloroplast genome was conducted using Geneious Prime v2020.2.4 (Biomatters Ltd., Auckland, New Zealand) based on the previously reported Ericaceae chloroplast genomes in the National Center for Biotechnology Information (NCBI) database. tRNA prediction was performed using the tRNAscan-SE2.0 (Chan and Lowe, 2019), and a circular map was drawn using OGDRAW v1.31 (Greiner et al., 2019).
The complete chloroplast genome sequences of 15 Rhododendron species were downloaded from GenBank (https://www.ncbi.nlm.nih.gov/genbank/) to investigate the phylogenetic relationship of R. caucasicum with other Rhododendrons. Among the previously reported complete chloroplast genomes from Ericaceae species, Gaultheria longibracteolata R.C. Fang and Vaccinium myrtillus L. were used as the outgroups. Phylogenetic analysis was performed using 74 coding sequences of Rhododendron species. Alignments were performed using Clustal Omega v1.2.2 as implemented in Geneious Prime software, and the alignments were concatenated. Subsequent phylogenetic analyses hereafter were performed in PhyloSuite v1.2.3 (Zhang et al., 2020; Xiang et al., 2023). The optimal partitioning strategies and evolutionary models for the coding sequences under the Bayesian information criterion were determined using ModelFinder (Kalyaanamoorthy et al., 2017). The best-fit partition models are shown in Table 2. A maximum likelihood (ML) was reconstructed using IQ-tree (Nguyen et al., 2015) with 10,000 ultrafast bootstrap replicates (Minh et al., 2013). A Bayesian inference tree was built using MrBayes v3.2.7a (Ronquist et al., 2012). Markov Chain Monte Carlo runs were performed for 10 million generations, and trees were sampled every 1,000 generations. The first 25% of the trees were discarded as burn-in to ensure the chains were stationary. The remaining trees were used to generate a strict consensus tree and calculate each node’s posterior probabilities.
RESULTS AND DISCUSSIONThe chloroplast genome of R. caucasicum (GenBank accession no. OQ998973) consists of 199,487 bp and has four subregions: a large single-copy region (LSC) of 107,645 bp and a small single-copy region (SSC) of 2,598 bp that are separated by the inverted repeat regions (IR) of 44,622 bp (Fig. 1). The chloroplast genome’s GC content is 35.9% and is 35.3, 30.0, and 36.7% in the LSC, SSC, and each of the IRs, respectively. The chloroplast contains 143 genes (93 protein-coding genes [PCGs], eight ribosomal RNAs [rRNAs], and 42 transfer RNAs [tRNA]); 24 genes (13 PCGs, four rRNAs, and nine tRNAs) are duplicated in the IR regions (Table 1). clpP, ycf2, and ycf68 were not identified in the R. caucasicum cp genome, and we concluded those genes were missing since they were also missing in the previously reported Rhododendron cp genomes (Liu et al., 2020; Ma et al., 2021; Wang et al., 2021).
The R. caucasicum chloroplast genome size (199,487 bp) falls within the known size categories of Rhododendron genomes, ranging from 197,877 bp (R. mole; MZ073672) to 230,777 bp (R. kawakamii, NC058233), which is relatively large among the angiosperm chloroplast genomes (Daniell et al., 2016; Olejniczak et al., 2016). The R. caucasicum chloroplast genome has expanded IRs and contracted SSC like other previously reported Rhododendron cp genomes. nhhA, ndhD, ndhE, ndhG, ndhH, ndhI, rps15, psaC, ccsA, and rpl32, which are generally found in the SSC, were moved to the IR, while only ndhF was detected in the SSC region of R. caucasicum. Thus, the increased chloroplast genome size might be due to the expansion of the IRs.
The ML- and Bayesian inference-based phylogenies had the same topology with high support for each branch (Fig. 2). The sub-generic relationships shown in this study are consistent with previous molecular phylogenetic studies (Shrestha et al., 2018; Xia et al., 2022). Except for the subgenus Therorhodion, which is not included in this study, two species in the subgenus Tsutsui diverged first from the rest. Then, the subgenus Rhododendron diverged from the subgenera Hymenanthes and Pentanthera.
Given that R. caucasicum is a tertiary relic species and the closest sister to R. aureum and R. brachycarpum (Milne, 2004), we expect that further extensive phylobiogeographic studies will clarify their speciation histories and provide clues to their disjunct distribution. Accordingly, the chloroplast sequence we describe of R. caucasicum will provide useful information for future studies to understand their phylogenetic and evolutionary relationships.
ACKNOWLEDGMENTSThis research was supported by grants from the National Institute of Biological Resources, funded by the Ministry of Environment of the Republic of Korea (Grant No. NIBR202207101). This project was carried out in collaboration under the Memorandum of Understanding signed by National Institute of Biological Resources and Ilia State University. The authors are grateful to Prof. Ohseok Kwon at Kyungpook National University for his work on this cooperative project and to Dr. Jongsun Park and Dr. Woochan Kwon at Infoboss for their assistance on assembly and annotation.
Fig. 1.Circular map of the Rhododendron caucasicum complete chloroplast genome. The genes outside the circle are transcribed clockwise while those inside are transcribed counterclockwise. The dark gray plot in the inner circle corresponds to the GC content. Large single-copy, small single-copy, and inverted repeat are indicated by LSC, SSC, and IR (IRA and IRB), respectively. 
Fig. 2.Phylogenetic tree of Rhododendron caucasicum and related taxa based on 74 protein-coding gene sequences of the chloroplast sequence. The phylogenetic tree was drawn based on the maximum likelihood phylogenetic tree. The number above the branches corresponds to the bootstrap support values from the maximum likelihood and posterior probability values for the Bayesian inference analyses. Gaultheria longibracteolata and Vaccinium myrtillus were used as outgroups. The numbers in parenthesis are National Center for Biotechnology Information (NCBI) GenBank accession numbers. 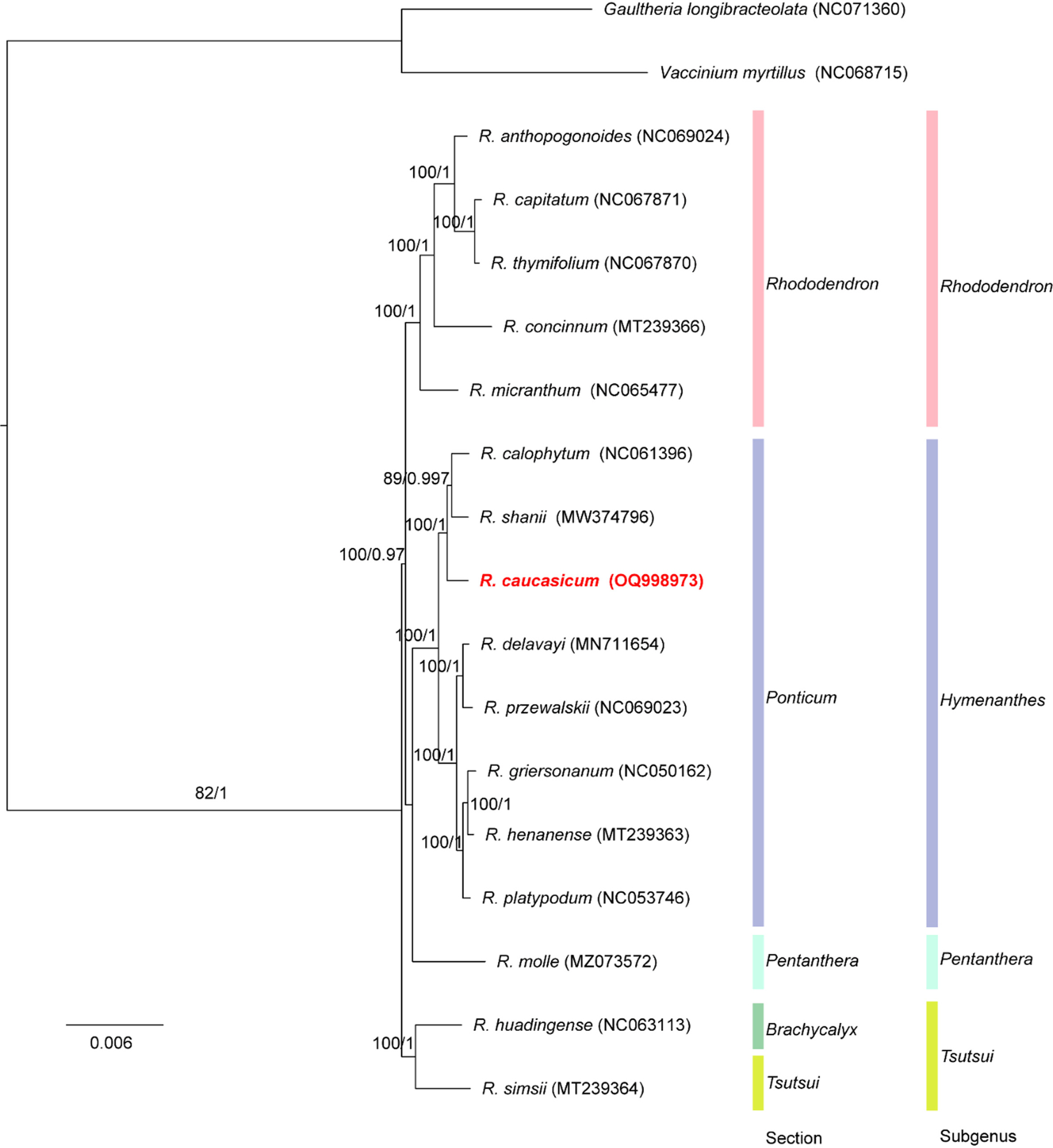
Table 1.List of genes annotated in the chloroplast genome of Rhododendron caucasicum.
Table 2.Optimal partitioning strategies and evolutionary models selected by ModelFinder using the Bayesian information criterion. ML, maximum likelihood; BI, Bayesian information; TVM, transversion model, AG = CT and unequal base frequency; TIM3, transition model, AC = CG, AT = GT and unequal base frequency; TPM2u, AC = AT, AG = CT, CG = GT and unequal base frequency; TPM3u, AC = CG, AG = CT, AT = GT and unequal base frequency; F81, equal rates but unequal base frequency; GTR, general time reversible model with unequal rates and unequal base frequency; HKY, unequal transition/transversion rates and unequal base frequency; F, empirical base frequency; G4, discrete gamma model with four rate categories; I, allowing for a proportion of invariable sites; R2, freerate model parameters with two of categories; R3, freerate model parameters with tree of categories. LITERATURE CITEDBolger, A.M. Lohse, L and Usadel, B. 2014. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114-2120.
Chan, P. P and Lowe, T. M. 2019. tRNAscan-SE: Searching for tRNA genes in genomic sequences. Gene Prediction. Methods in Molecular Biology. 1962: Kollmar, M (ed.), Humana, New York. 1-14.
Daniell, H. Lin, C.-S. Yu, M and Chang, W.-J. 2016. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biology 17: 134.
Doyle, J. J and Doyle, J. L. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19: 11-15.
Fan, Y. Jin, Y. Ding, M. Tang, Y. Cheng, J. Zhang, K and Zhou, M. 2021. The complete chloroplast genome sequences of eight Fagopyrum species: Insights into genome evolution and phylogenetic relationships. Frontiers in Plant Science 12: 799904.
Gitzendanner, M. A. Soltis, P. S. Wong, G. K.-S. Ruhfel, B. R and Soltis, D. E. 2018. Plastid phylogenomic analysis of green plants: A billion years of evolutionary history. American Journal of Botany 105: 291-301.
Greiner, S. Lehwark, P and Bock, R. 2019. OrganellarGenome-DRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Research 47: W59-W64.
Kalyaanamoorthy, S. Minh, B. Q. Wong, T. K. F. von Haeseler, A and Jermiin, L. S. 2017. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nature Methods 14: 587-589.
Kawabe, A. Nukii, H and Furihata, H. Y. 2018. Exploring the history of chloroplast capture in Arabis using whole chloroplast genome sequencing. International Journal of Molecular Sciences 19: 602.
Li, H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at arXiv
https://.org/10.48550/arXiv.13033997.
Li, H.-T. Yi, T.-S. Gao, L.-M. Ma, P.-F. Zhang, T. Yang, J.-B. Gitzendanner, M. A. Fritsch, J. Cai, P. W. Luo, Y. Wang, H. van der Bank, M. Zhang, S.-D. Wang, Q.-F. Wang, J. Zhang, Z.-R. Fu, C.-N. Yang, J. Hollingsworth, P. M. Chase, M. W. Soltis, D. E. Soltis, P. S and Li, D.-Z. 2019. Origin of angiosperms and the puzzle of the Jurassic gap. Nature Plants 5: 461-470.
Liu, B.-B. Ren, C. Kwak, M. Hodel, R. G. J. Xu, C. He, J. Zhou, W.-B. Huang, C.-H. Ma, H. Qian, G.-Z. Hong, D.-Y and Wen, J. 2022. Phylogenomic conflict analyses in the apple genus Malus s.l. reveal widespread hybridization and allopolyploidy driving diversification, with insights into the complex biogeographic history in the Northern Hemisphere. Journal of Integrative Plant Biology 64: 1020-1043.
Ma, L.-H. Zhu, H.-X. Wang, C.-Y. Li, M.-Y and Wang, H.-Y. 2021. The complete chloroplast genome of Rhododendron platypodum (Ericaceae): An endemic and endangered species from China. Mitochondrial DNA Part B: Resources 6: 196-197.
Milne, R.I. 2004. Phylogeny and biogeography of Rhododendron subsection Pontica, a group with a tertiary relic distribution. Molecular Phylogenetics and Evolution 33: 389-401.
Milne, R. I. Davies, C. Prickett, R. Inns, L. H and Chamberlain, D. F. 2010. Phylogeny of Rhododendron subgenus Hymenanthes based on chloroplast DNA markers: Between-lineage hybridization during adaptive radiation? Plant Systematics and Evolution 285: 233-244.
Minh, B. Q. Nguyen, M. A. T and von Haeseler, A. 2013. Ultrafast approximation for phylogenetic bootstrap. Molecular Biology and Evolution 30: 1188-1195.
Nguyen, L.-T. Schmidt, H. A. von Haeseler, A and Minh, B. Q. 2015. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32: 268-274.
Olejniczak, S.A. Łojewska, E. Kowalczyk, T and Sakowicz, T. 2016. Chloroplasts: State of research and practical applications of plastome sequencing. Planta 244: 517-527.
Ronquist, F. Teslenko, M. van der Mark, P. Ayres, D. L. Darling, A. Höhna, S. Larget, B. Liu, L. Suchard, M. A and Huelsenbeck, J. P. 2012. MrBayes 32: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539-542.
Shrestha, N. Wang, Z. Su, X. Xu, X. Lyu, L. Liu, Y. Dimitrov, D. Kennedy, J. D. Wang, Q. Tang, Z and Feng, X. 2018. Global patterns of Rhododendron diversity: The role of evolutionary time and diversification rates. Global Ecology and Biogeography 27: 913-924.
Wang, Z.-F. Chang, L.-W and Cao, H.-L. 2021. The complete chloroplast genome of Rhododendron kawakamii (Ericaceae). Mitochondrial DNA Part B: Resources 6: 2538-2540.
Xia, X.-M. Yang, M.-Q. Li, C.-L. Huang, S.-X. Jin, W.-T. Shen, T.-T. Wang, F. Li, X.-H. Yoichi, W. Zhang, L.-H. Zheng, Y.-R and Wang, X.-Q. 2022. Spatiotemporal evolution of the global species diversity of Rhododendron
. Molecular Biology and Evolution 39(1): msab314.
Xiang, C.-Y. Gao, F. Jakovlić, I. Lei, H.-P. Hu, Y. Zhang, H. Zou, H. Wang, G.-T and Zhang, D. 2023. Using PhyloSuite for molecular phylogeny and tree?based analyses. iMeta 2: e87.
Zerbino, D.R and Birney, E. 2008. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Research 18: 821-829.
|
|
|||||||||||||||||||||||||||||||||||