DNA barcoding is a molecular technique which is used to identify a specimen with DNA sequences of short regions (Hebert et al., 2003; Kress et al., 2005; Xiang et al., 2011). With the advent of DNA sequencing methods and the lower cost of handling samples, the DNA barcoding method has proved to be a novel tool for the conservation of biodiversity at the global (Hebert et al., 2004; CBOL Plant Working Group, 2009) and the local levels (Lahaye et al., 2008; Tripathi et al., 2013; Kim et al., 2014; Zhang et al., 2015). With regard to plants, the rbcL and matK regions in the chloroplast genome are widely utilized by various groups (e.g., CBOL Plant Working Group, 2009). These DNA barcode regions are useful for identifying unknown specimens into families and genera, and sometimes to the species level (Lahaye et al., 2008). Additional DNA barcoding regions in chloroplast regions, such as trnH-psbA, atpF-atpH, and psbK-psbI, and Internal Transcribed Spacers (ITS) in nuclear ribosomal DNA have been suggested to augment the resolution power to lower taxonomic levels (Tripathi et al., 2013; Kim et al., 2014).
Because of its advantage for the identification of species from a small fragment even without knowledge of the key morphological features and because a large number of samples can be processed in a short period of time, DNA barcoding has been particularly useful for non-specialists in plant taxonomy. Building a regional DNA barcode database for all known species is the first step in the making the DNA barcode method useful for diverse applications, such as trade control at the customs, conservation biology, the utilization of plant resources in bio-industries, and for forensic science. An evaluation of the utility of DNA barcode regions for each group of plants concerning the level of resolution of species given a set of DNA barcodes would provide a basic information for application (CBOL Plant Working Group, 2009).
Schisandraceae is a small family consisting of Schisandra Michx. with approximately 23 species and Kadsura Juss with ca. 22 species (Saunders, 1998, 2000). Plants of Schisandraceae are evergreen or deciduous woody vines with alternate and simple leaves and unisexual flowers with spirally arranged carpels or stamens. Members of this family are distributed in temperate and subtropical forests in eastern and Southeast Asia, except for S. glabra (Brickell) Rehder, which grows in the southeastern United States and central Mexico. In Korea, S. chinensis (Turcz.) Baill., S. repanda (Siebold & Zucc.) Radlk., and K. japonica (L.) Dunal are distributed. While the latter two species are restricted to southern areas within Korea, S. chinensis is widely distributed. Schisandra chinensis is also cultivated for tonic beverages and oriental medicine (Panossian and Wikman, 2008; Jang et al., 2016).
Leaf and floral characters are important when classifying Schisandraceae (Saunders, 1998, 2000; Suh, 2007). When a specimen has a relatively complete morphology, each species is easily distinguished from one another. However, when only part of the plant is available for identification, it is difficult to classify a species with the morphological features. Furthermore, the three aforementioned species are distributed in neighboring countries. Schisandra chinensis is also distrubited in China, Russia, and Japan, and S. repanda and K. japonica also occur in Japan (Ohwi, 1965; Saunders, 1998, 2000). Lee et al. (2013) have developed species-specific RAPD (Random Amplified Polymorphic DNA) markers and SCAR (Sequence Characterized Amplified Region) markers for the Korean Schisandraceae and the Chinese S. sphenanthera Rehder & E. H. Wilson. These molecular markers would be very useful to authenticate the specific species, in which unknown materials are one of the four species. DNA barcoding method would provide an additional molecular tool to identify a species, which does not require a prior knowledge about the species. The establishment of a regional DNA barcode database for the Korean population and an assessment of the utility of DNA barcoding for geographic structures are necessary to make the DNA barcoding tool useful (Kim et al., 2014). Recently, DNA barcoding loci were evaluated for Schisandraceae (Zhang et al., 2015), focusing on the development of DNA barcode markers for the Chinese populations of Schisandraceae but not on evaluating the level of molecular variation across the geographic ranges of widely distributed species.
The main objectives of the present study are (1) to develop a DNA barcode database for Korean Schisandraceae, (2) to assess the level of differentiation of the species, and (3) to evaluate the utility of DNA barcodes.
Materials and Methods
Plant materials
Thirteen samples of Schisandraceae in total were included in this study (Table 1). For the relatively common and widespread species of S. chinensis, seven plants were included to represent their distributional range. For K. japonica, restricted to the southern part of Korea, five plants were analyzed. Schisandra repanda is very rare, and one sample was included in this study. Voucher specimens were deposited at the Herbarium of the National Institute of Biological Resources (KB). We included Illicium anisatum L. as an outgroup in the phylogenetic analysis based on phylogenetic analyses (Hao et al., 2001; Liu et al., 2006; Zhang et al., 2015).
DNA extraction, amplification, and sequencing
Total DNA was extracted from fresh leaves or dried leaf materials with silica gel using the DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Two DNA barcode regions of chloroplast DNA (cpDNA), the rbcL and matK regions, and one nuclear region (ITS) were amplified via polymerase chain reactions (PCR). Primers for the cpDNA barcodes were published in previous studies (Soltis et al., 1992; Sang et al., 1997; Cuénoud et al., 2002; Kress and Erickson, 2007) and are summarized in Kim et al. (2014). The ITS region was amplified using the primers its6 (5′–CCG CTT ATT GAT ATG CTT AAA CT–3′) and its9 (5′–TCG TAA CAA GGT TTC CGT AGG TG–3′). The new primers for the ITS region, developed by the last author, have been shown to be useful to amplify difficult samples (Potter et al., 2007, Daniel et al., 2008). For each PCR reaction, 1 μL of total DNA was included in a 20 μL reaction mixture with Solg EF-Taq DNA polymerase (Solgent, Daejeon, Korea). Amplification of the target regions was conducted with a Veriti thermal cycler (Applied Biosystems, Carlsbad, CA, USA) under the following conditions: initial denaturation at 95°C for 2 min, 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min, followed by a final extension at 72°C for 5 min. PCR products were examined on a 1% agarose gel in 1× TBE buffer, purified and were sent to Solgent for sequencing, which prepared the sequencing reaction using the same primers used in PCR with the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). Sequences were determined using the 3730xl DNA analyzer (Applied Biosystems).
Data analysis
Sequences were edited in Sequencher version 5.0 (Gene Codes Corporation, Ann Arbor, MI, USA), aligned using MUSCLE (Edgar, 2004), and adjusted manually as needed. The sequence data generated in this study, designated as the Korean Schisandraceae data, were analyzed separately and in combination with previously published data which contains DNA barcode sequences from Chinese and Japanese samples (Zhang et al., 2015). In their study, Zhang et al. (2015) included 14 species of Schisandra and six of Kadsura, including S. chinensis, S. repanda, and K. japonica. We obtained DNA sequences of rbcL, matK, and the ITS regions of all accessions of the three species published in Zhang et al. (2015) and analyzed these together with our data to examine whether the Korean accessions can be distinguished from the Chinese and Japanese accessions by the DNA barcodes. In addition, exemplar accessions of other species were included in a more inclusive analysis (Appendix 1). The expanded data included 36 accessions from 13 species of Schisandra and 13 from five of Kadsura. Phylogenetic analyses were conducted using the maximum parsimony (MP) and neighbor-joining methods (NJ) in PAUP* 4.0b10 (Swofford, 2002). All characters were treated as unordered and were weighted equally in the MP analyses. Gaps resulting from multiple alignments of indels were treated as missing. Heuristic searches were used with 100 replicates of random sequence additions with tree bisection-reconnection branch swapping, with all of the best trees saved at each step (MulTrees). Kimura’s two-parameter model was employed to calculate the distance matrix in NJ analyses. Bootstrap analyses (Felsenstein, 1985) of 500 pseudoreplicates were conducted using both MP and NJ criteria. For the bootstrap analysis using the MP method, heuristic searches were employed with a simple sequence addition in PAUP* to evaluate the support for the Korean Schisandraceae data. For the expanded data matrix, 10,000 pseudoreplicates were sampled using the “fast” stepwise-addition option in PAUP*.
The resolution ability of the species for each DNA barcode and a combinatory DNA barcode were examined based on the percentage of monophyletic species given the MP and NJ trees. When only one accession in a species was included, it was treated as monophyletic if the branch leading to the accession was greater than zero.
Results and Discussion
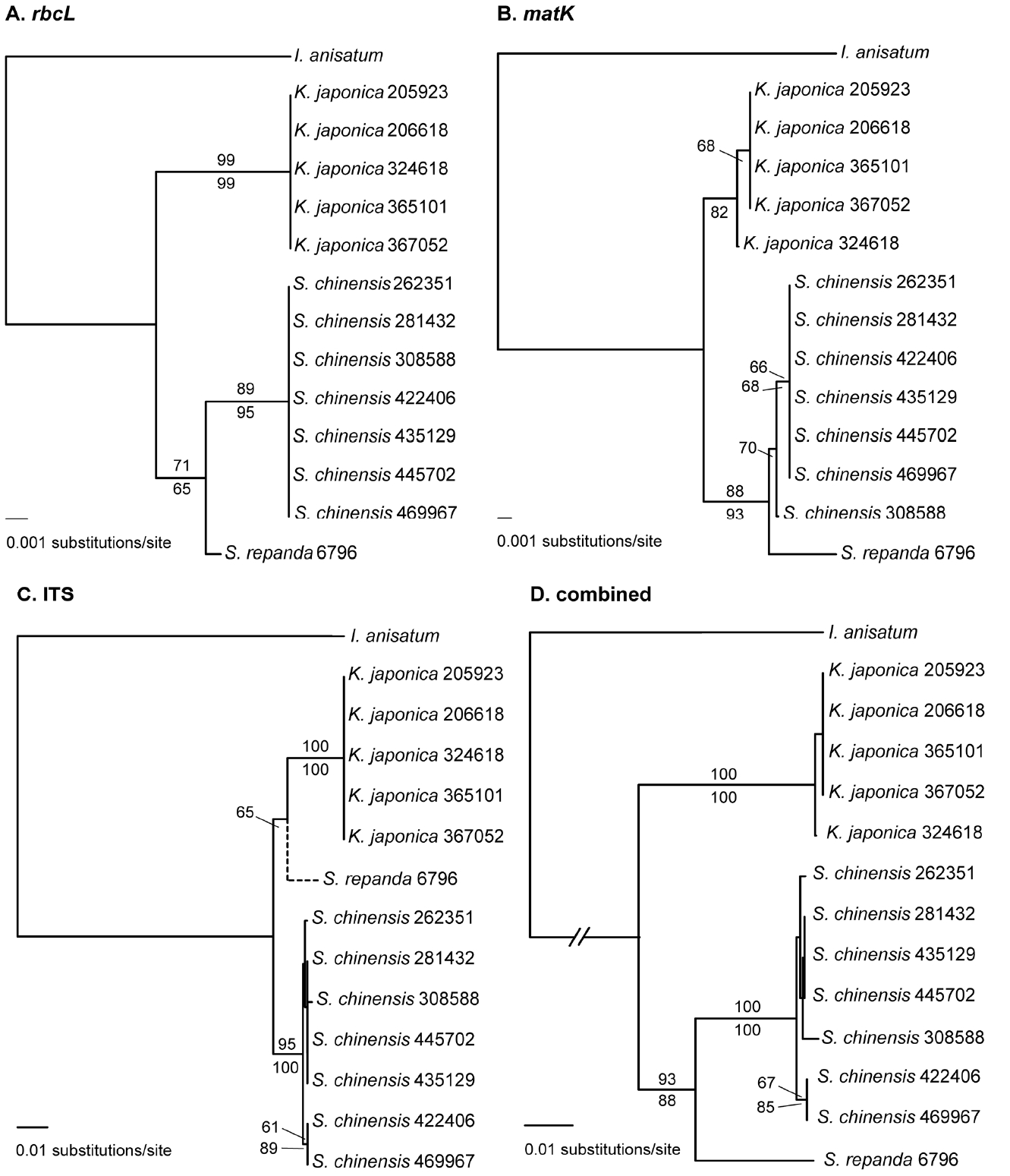
The primers for the amplification and sequencing of the rbcL, matK, and ITS regions for the Korean Schisandraceae worked well, producing a success rate of 100%. The lengths of the amplified rbcL and matK regions were 646 bp and 867 bp, respectively. The lengths of the ITS regions were 669 bp in S. repanda, 670 bp in S. chinensis, and 672 bp in K. japonica. The size within a species was invariable in all three regions. The rbcL sequences of all accessions of S. chinensis included were identical, as they were for K. japonica. In the matK data, five haplotypes were found, two in S. chinensis, two in K. japonica, and one in S. repanda. The ITS data produced intraspecific variation only in S. chinensis, in which four ribotypes were found among seven accessions. The final alignment of the concatenated data with Illicium anisatum as an outgroup resulted in 2,274 sites, of which 194 sites were variable and 44 were parsimoniously informative (Table 2).
NJ analyses of the three regions, separately and simultaneously, suggested that all three species were distinguishable by DNA barcodes (Fig. 1). MP analyses produced the same results as NJ analyses, except for the separate analysis from matK, in which relationship between S. chinensis and S. repanda was not resolved (Table 2). In all NJ and MP analyses, except for MP analysis of matK, multiple accessions of S. chinensis and K. japonica formed monophyletic groups with strong bootstrap values. Schisandra repanda, where only one accession was included, was divergent from S. chinensis and K. japonica and was not nested within either of the two species. Schisandra repanda was a sister to S. chinensis in the rbcL and matK data with relatively high support values, resulting in mutually exclusive clades of Schisandra and Kadsura (Fig. 1A, B). Schisandra repanda, however, was resolved as a sister to K. japonica in the NJ analysis of the ITS data with a bootstrap value of 65% (Fig. 1C). The NJ tree is the only topology that groups S. repanda with K. japonica. The MP analysis of the ITS data suggested that S. repanda is a sister to S. chinensis. A combined analysis of the three regions showed that S. repanda is a sister to S. chinensis in both the NJ and MP methods. Our data indicate that all of the DNA barcode regions included in this study (rbcL, matK, and ITS) are useful for identifying a species of Schisandraceae in Korea. The data also suggest that the resolution ability to distinguish all of the Korean species may be obtained by a single DNA barcode and a combinatorial barcode.
Our DNA barcode data are consistent with the morphology in that the three species of Schisandraceae in Korea are morphologically distinct (Saunders, 1998, 2000; Suh, 2007). Kadsura japonica is easily distinguished from S. chinensis and S. repanda by having globose aggregate of berries. Fruits of S. chinensis and S. repanda are aggregate berries that are loosely arranged along the axis, resembling the racemose infructescence, as the receptacle elongates when the fruit is mature. Plants of K. japonica are evergreen, while those of S. chinensis and S. repanda are deciduous. Schisandra repanda is rare in Korea, only distributed at high elevations on Mt. Hallasan in Jeju-do. This species can be distinguished from S. chinensis by having ovate or broadly elliptic leaves and black fruits. Stamens S. repanda are arranged at the margin of the flat pentagonal synandrous appendage, whereas those of S. chinensis are attached onto the columnar receptacle (Saunders, 2000; Liu et al., 2006).
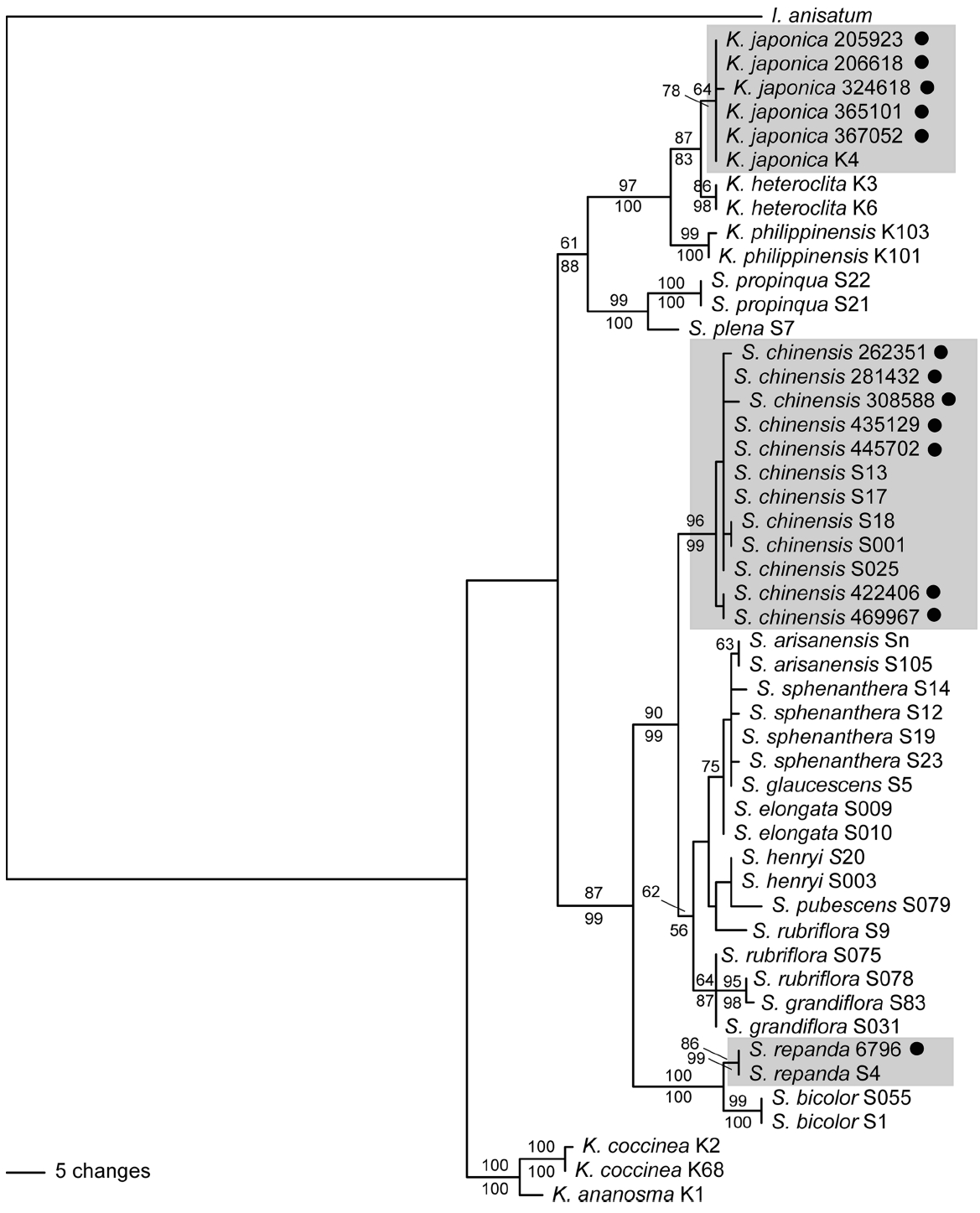
Distinctiveness of the three species of Schisandraceae (S. chinensis, S. repanda, and K. japonica) was also supported at the species level when Chinese and Japanese populations were added to the combined data (Fig. 2). Kadsura japonica was a sister to K. heteroclita (Roxb.) Craib and S. repanda was a sister to S. bicolor W. C. Cheng. Schisandra chinensis is a sister to the clade of S. arisanensis Hayata and S. grandiflora Hook. f. & Thompson (Fig. 2). Among the three regions, ITS regions provide the best resolution power in the expanded data (Table 2). In the rbcL data, only S. chinensis was resolved as monophyletic in the MP analysis, while S. chinensis and K. japonica were resolved as monophyletic in the NJ analysis. The matK data alone did not have sufficient power to resolve the three species in the expanded data (Table 2). Therefore, using three concatenated regions is the best option for DNA barcoding in Schisandraceae in Korea. The combined analyses of the expanded data showed that both Schisandra and Kadsura were not monophyletic (Fig. 2). Previous molecular phylogenetic studies of Schisandraceae also produced similar results (Hao et al., 2001; Liu et al., 2006; Zhang et al., 2015). Further detailed systematic studies are needed to investigate the generic boundaries of the two genera.
There is a clear gap between the distribution of the intraspecific distance and the distribution of the interspecific distance in all three species and in the combined data (Fig. 3). The minimum value of the interspecific distances among S. chinensis, S. repanda, and K. japonica in the DNA barcode regions either separately or in combination are 4 to 23 times higher than the maximum value of the intraspecific distance in the three species (Table 2). These results indicate that each species is genetically well differentiated and that all of DNA barcodes employed in this study should have power to distinguish a species.
Our data showed that the three DNA barcoding regions do not resolve the geographic origins of the samples in S. chinensis. Seven individuals of S. chinensis as sampled here were intended to represent the complete distributional range in Korea. The five Chinese samples used in Zhang et al. (2015) were collected in the Beijing and Jilin areas. We found four sequence types in Korea and two in China. Of these, one of the types in Korea with three populations (accession Nos. 281432, 435129, and 445702) was identical to a type in China with three individuals (accession Nos. S13, S17, and S025) in the combined data. The three Korean accessions were collected in montane areas of eastern Korea, such as Gangwon-do and Pohang-si (Table 1). The shared sequence type between Korea and China may represent a widely distributed type, as the sequence type was predominantly found in six out of twelve individuals. The other three sequence types derived from four individuals in Korea are unique, distinct from the second sequence type of China (accession Nos. S001 and S18). These Korean types may represent local types which are differentiated from common and ancestral types. However, S. chinensis is widely distributed in northeastern China, Mongolia, far eastern Russia, Korea, and Japan. Further studies are necessary to determine the geographic structure of the species with more samples of all distributional ranges and rapidly evolving molecular markers.
Schisandra repanda distributed in Korea and Japan was supported as a monophyletic group, a sister to S. bicolor (Fig. 2). This suggests that S. repanda is a well-supported species and that the DNA barcode regions have sufficient power to identify the species. Our study and that of Zhang et al. (2015) only sampled one individual of S. repanda. The sequences of the combined regions were identical in the two accessions, suggesting that the Korean population is undifferentiated from the Japanese one. These results also suggest that molecular marker for fine-scale analysis, such as microsatellite markers, should be employed to identify the geographic origins of samples in S. repanda.
Kadsura japonica is distributed in southern Korea and Japan (Honshu, Shikoku, Kyushu, and on the Ryukyu Islands) (Ohwi, 1965; Walker, 1976). Samples of K. japonica analyzed in this study were all from Korea, as was the sample used in Zhang et al. (2015). Our data indicate that the DNA barcode regions have sufficient resolution power to identify K. japonica. There are two sequence types within K. japonica. Accession No. 324618 from Jejudo Island differed from the remaining accessions by one base in the matK region. All other samples including those from Jejudo Island produced identical sequences. This suggests that the degree of genetic diversity in K. japonica in Korea is low.