INTRODUCTION
Scrophularia L. belongs to the family Scrophulariaceae, which comprises approximately 200–270 species (Ortega Olivencia, 2009; Jang et al., 2021). This genus is distributed throughout the Northern Hemisphere and has square stems (sometimes winged stems) and generally opposite leaves. Stiefelhagen (1910) divided this genus into two sections, sect. Anastomosantes Stiefelhagen (sect. Scrophularia) and sect. Scorodoniae (Benth.) Stiefelhagen (sect. Canina) which are characterized by perennial subshrubs, petal length, corolla tube shape, and life form. The two series in sect. Scrophularia are Grayanaea T. Yamaz and Kakudenses. T. Yamaz recognized (Yamazaki, 1949).
Seven species of Scrophularia (S. buergeriana, S. koraiensis, S. kakudensis, S. takesimensis, S. cephalantha, S. kakudensis var. microphylla, and S. alata) are distributed throughout Korea (Jang and Oh, 2013). All Korean Scrophularia species are included in sect. Scrophularia. Two species (S. alata and S. takesimensis) belong to the series Grayanae and five species (S. buergeriana, S. kakudensis, S. kakudensis var. microphylla, S. koraiensis, and S. cephalantha) belong to series Kakudenses. However, identifying S. kakudensis is difficult due to similar morphological characteristics among S. kakudensis var. microphylla, S. cephalantha, and S. pilosa. Jang et al. (2011) suggested that S. pilosa should be treated as a synonym for S. kakudensis. Scrophularia cephalantha and S. kakudensis var. microphylla are distinct from S. kakudensis during the flowering season, with fewer nodes numbers on the stem and the sizes of the leaves, respectively. Recently, the phylogenetic relationships and evolution of several Scrophularia species have been studied using chloroplast (cp) genomes (Xu et al., 2018; Jang et al., 2021; Wang et al., 2022). In this study, we sequenced and analyzed the cp genome of S. kakudensis and performed a comparative analysis of S. kakudensis and S. cephalantha.
MATERIALS AND METHODS
We collected young leaves of S. kakudensis from Mt. Bohyeonsan (36.16625°N, 128.98097°E). The voucher specimens were deposited at the Daegu National Science Museum. Total DNA was extracted using the DNeasy Plant Mini Kit (QIAGEN Inc., Valencia, CA, USA). Genomic DNA was sequenced using the Illumina HiSeq X platform (San Diego, CA, USA). We obtained 37,322,320 reads from the 150 bp paired-end sequences. The chloroplast genomes were assembled using GetOrganelle (Jin et al., 2020) and Geseq (Tillich et al., 2017) and Geneious Prime v.2022.1.1 (http://www.geneious.com) were used to annotate the S. kakudensis cp genome. Chloroplast genome mapping was performed using OGDRAW v. 1.3.1 (Greiner et al., 2019).
The chloroplast genome sequences of six taxa (Table 1), including one outgroup (Verbascum phoenieum, MN983301), were included in the phylogenetic analyses. The 78 protein-coding genes shared across taxa were extracted from each chloroplast genome and concatenated. The sequences were aligned using MAFFT (Katoh et al., 2022), and ML analysis was conducted using RAxML (Stamatakis, 2014) with the GTR + GAMMA + I model (rapid bootstrap of 1,000 replications). The complete cp genomes of S. kakudensis and S. cephalantha were compared. The nucleotide diversity (Pi) was determined using DnaSP (Rozas et al., 2017). The step size was set to 200 bp, and the window length was set to 600 bp.
RESULTS AND DISCUSSION
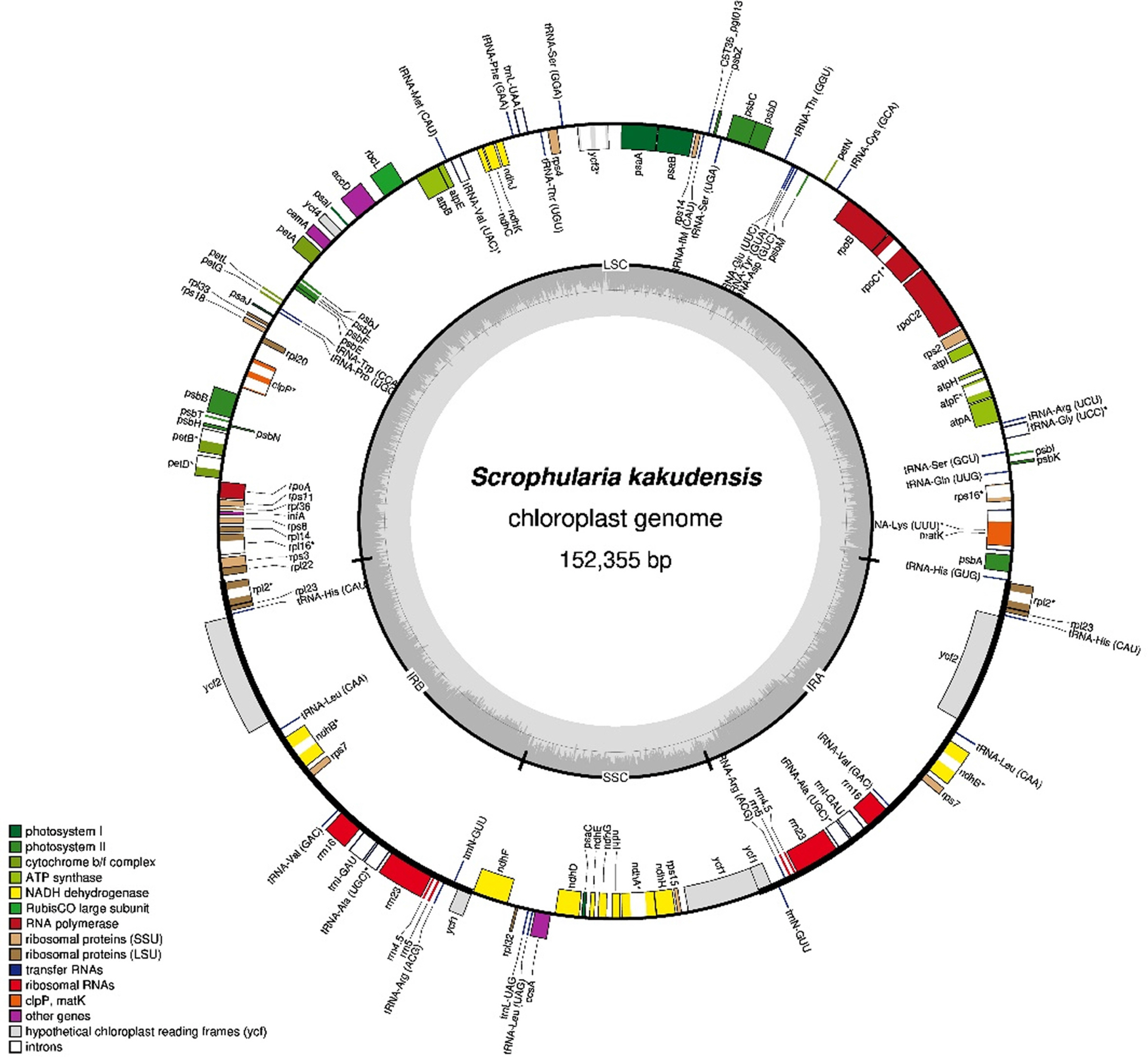
The complete cp genome of S. kakudensis (NCBI accession number: OR004236) comprised 152,355 bp with a quadripartite structure and two inverted repeat regions (IRs, 25,485 bp) separated by large single-copy (LSC, 83,476 bp) and small single-copy (SSC, 17,909 bp) regions (Fig. 1). The average GC content was 38.0%. It contained 112 genes, including 78 protein-coding, 30 tRNA, and 4 rRNA genes. Six protein-coding genes (rpl2, rpl23, ycf2, ndhB, rps7, and rps12), seven tRNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC) and four rRNA genes (4.5S, 5S, 16S, and 23 rRNA) were duplicated in two IR regions. Fifteen genes had one intron, and three genes (rps12, clpP, and ycf3) had two introns.
Comparative genomic analyses of Scrophularia revealed that the six cp genomes were highly conserved (Table 1). The total length of the Scrophularia species ranged from 152,355 bp (S. kakudensis) to 153,175 bp (S. ningpoensis). The GC content ranged from 38% to 38.1% (S. takesimensis), and the seven Scrophularia species contained the same genes (CDS, tRNA, and rRNA).
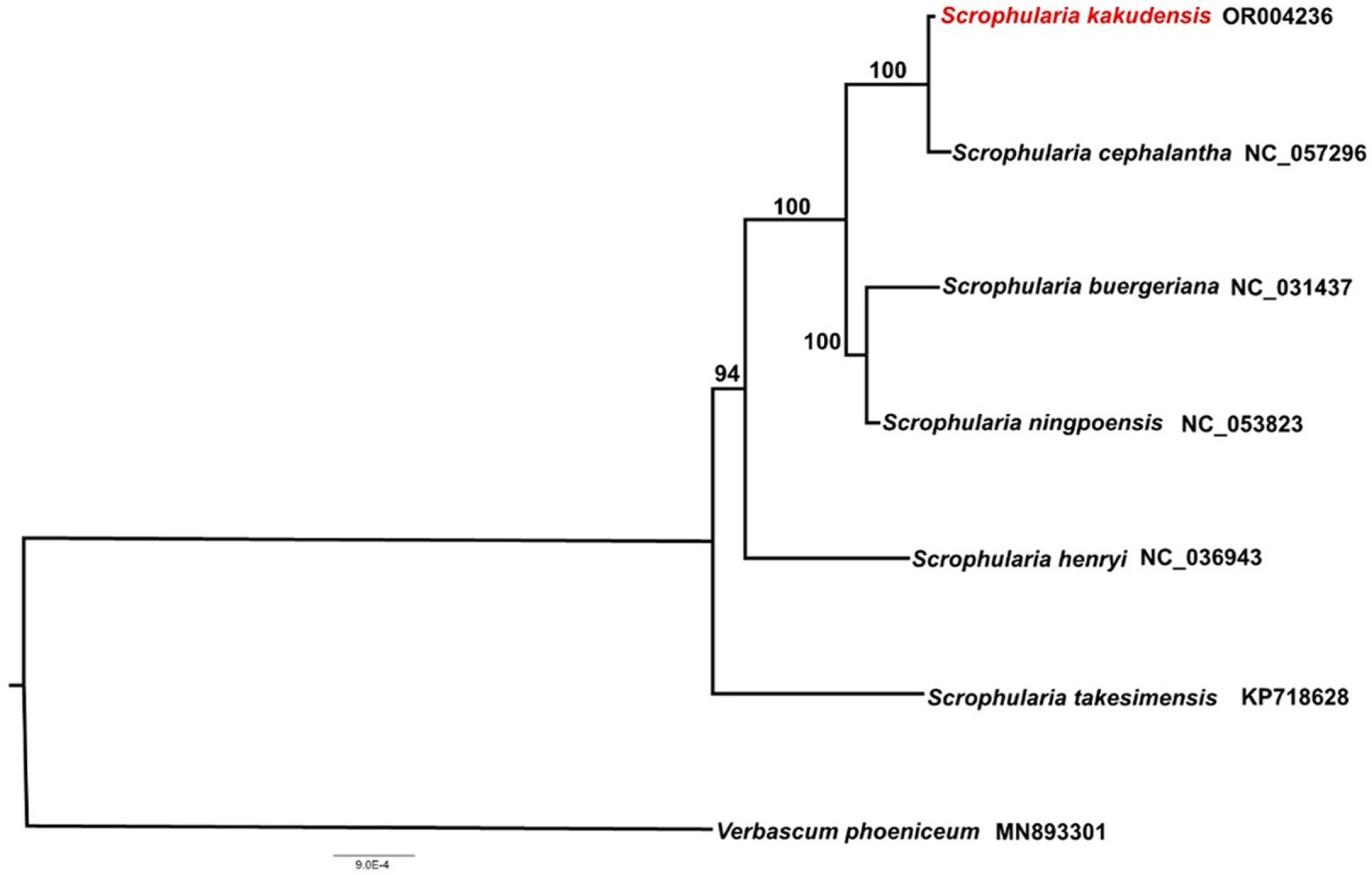
A phylogenetic analysis was conducted using 78 protein-coding genes from seven Scrophularia species and one outgroup (Verbascum phoeniceum). The genus Scrophularia is a monophyletic group. S. kakudensis was closely related to S. cephalantha with a 100% bootstrap value, and this clade was a sister to the S. buergeriana + S. ningpoensis clade (Fig. 2). This result supports the hypothesis that S. kakudensis and S. cephalantha are included in series Kakudenses (Yamazaki, 1949; Jang and Oh, 2013). Scrophularia cephalantha is an endemic species (Chung et al., 2023), and this study confirmed that it shows a close relationship between S. kakudensis and S. cephalatha.
Internal transcribed spacer (ITS) and cpDNA non-coding regions have been widely used to investigate molecular phylogeny at the interspecific level (Taberlet et al., 1991; Baldwin, 1992). In Scrophularia, Scheunert and Heubl (2010) tested the nrDNA (ITS) and chloroplast DNA intergenic spacers (psbA/trnH and trnQ/rps16). However, the relationship among Scrophularia species is not well supported. Recently, cp genome sequences have been used as genetic markers for DNA barcoding and phylogenetic relationships, and several studies on the genus Scrophularia have analyzed the chloroplast genome (Choi and Park, 2016;Yi and Kim, 2016; Jang et al., 2021; Wang et al., 2022; Guo et al., 2023).
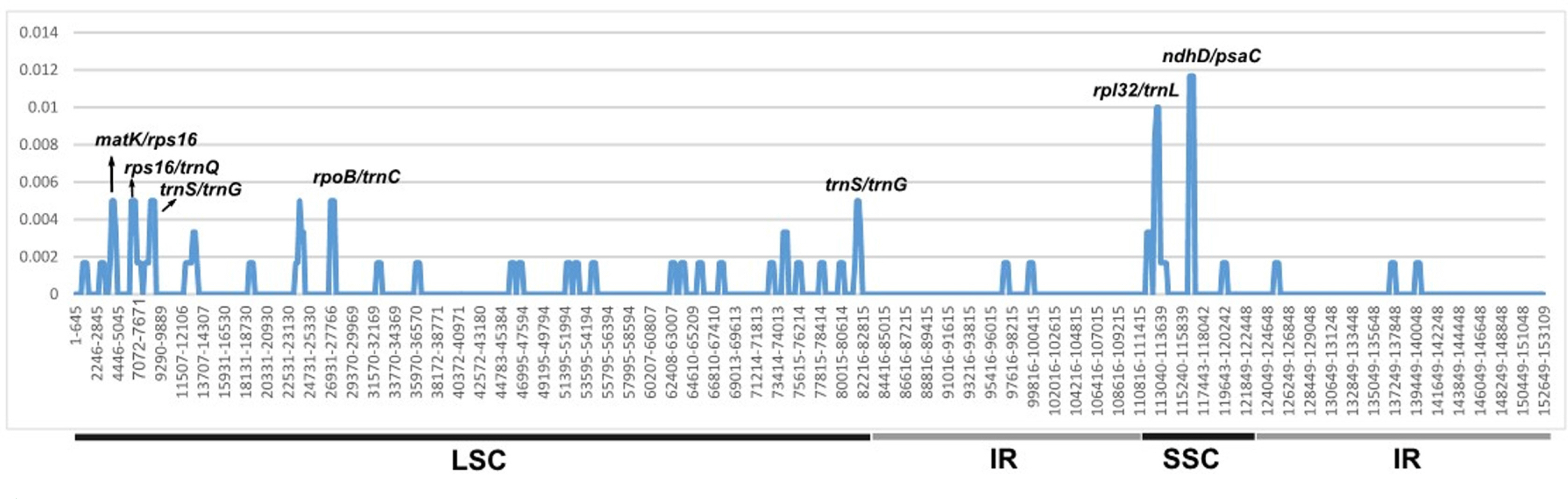
The nucleotide diversities between S. kakudensis and S. cephalantha were compared. Nucleotide diversity (Pi) ranged from 0–0.01167. Most of the variable regions were located between the ndhD/psaC regions (Pi = 0.01167). This result suggests that seven regions (matK/rps16, rps16/trnQ, trnS/trnG, rpoB/trnC, trnS/trnG, rpl32/trnL, and ndhD/psaC) were highly informative markers to identify species (Fig. 3).
Xu et al. (2018) suggested that nine markers (trnH/psbA, rps15, rps18/rpl20, rpl32/trnL, trnS/trnG, ycf15/trnL, rps4/trnT, ndhF/rpl32, and rps16/trnQ) could be used as DNA barcodes to distinguish Scrophularia. The results from this study indicate that, compared to the nine markers presented in the previous study, the rpl32/trnL and ndhD/psaC regions are more useful for studying S. kakudensis and its relatives. This study reports the complete chloroplast genome of S. kakudensis and provides useful information on the phylogenetic relationships within Scrophularia species.