INTRODUCTION
Rosaceae, commonly known as the rose family, contains more than 95 genera and 3,000 species (Potter et al., 2007; Hummer and Janick, 2009). In the family Rosaceae, the genus Spiraea contains more than 50 species and is mainly distributed in temperate and subtropical regions of the northern hemisphere (Hummer and Janick, 2009; Oh et al., 2010; Yu et al., 2018; Kostikova and Petrova, 2021). Spiraea prunifolia f. simpliciflora is a perennial shrub with ovate to oblonglanceolate leaves and an umbel inflorescence (Jang et al., 2020). This taxon is widely distributed in East Asia and is also cultivated in South Korea from the northern to southern parts of the country. It is primarily used for ornamental purposes, along with S. prunifolia Siebold & Zucc, which has double flowers (Jang et al., 2020). The Spiraea species are known to be useful horticultural and edible plants due to their beautiful flowers and high nectar content. The Spiraea species have traditionally been used as diuretics, antidotes, and painkillers in East Asia (Woo et al., 1996; Bae et al., 2012). Recently, it has also been reported that hydrothermal and ethanol extracts from the roots of S. prunifolia f. simpliciflora have antioxidant, anti-inflammatory, anti-cancer effects and that they protect nerve cells (Sim et al., 2017; Oh et al., 2018; Kim et al., 2019).
Plastid is an endosymbiotic organelle for photosynthesis that contains its own maternally inherited genome. In most plants, the plastid genome (plastome) is a circular molecule of 150–170 kb, generally consisting of a large single copy (LSC), a short single copy (SSC), and two inverted repeats (IR). The development of next-generation sequencing (NGS) has led to extensive studies of plastomes in land plants by reducing the time and cost required to assemble the plastome, with the conserved features and sufficient polymorphisms of the plastome enabling the plastome to be used for phylogenomic studies of various plants, such as Daphne and Viburnum (Leebens-Mack et al., 2005; Xu et al., 2015; Park et al., 2020; Yoo et al., 2021).
Along with the plastome data, the 45S and 5S nuclear ribosomal DNAs (nrDNAs) in the nuclear genome, which constitute the catalytic core of ribosomes, are also widely used for phylogenetic studies in land plants (Rodnina et al., 2007). The 45S nrDNA units are composed of three subunits (18S, 5.8S, and 26S rDNAs) and two internal transcribed spacer (ITS-1 and ITS-2) regions, and thousands of 45S units are tandemly repeated in the nuclear genome (Long and Dawid, 1980). Due to their structural advantage, the 45S and 5S rDNA sequences are highly conserved, making them a useful resource for phylogenetic studies of land plants (Lagesen et al., 2007). Previously, phylogenetic studies of the genus Spiraea using the nuclear ribosomal internal transcribed spacer region and a few plastid markers have revealed problems, such as discordance between different gene trees and polytomies due to insufficient informative sites for constructing phylogenies (Oh et al., 2010; Yu et al., 2018). In this study, we document the complete plastome and two nrDNA sequences of S. prunifolia f. simpliciflora. The results of our study can serve as a fundamental resource for further studies to understand these phylogenetic relationships and establish a classification of the genus Spiraea by considering phylogenetic evidence.
MATERIALS AND METHODS
Fresh leaves of S. prunifolia f. simpliciflora were collected from one plant identified at Mt. Gwanaksan in Seoul, South Korea. A voucher specimen was deposited in the National Institute of Biological Resources Herbarium (KB) under voucher number ZFTDVP0000000009. Total DNA was extracted from 100 mg of fresh leaves using the GeneAll Plant SV midi kit (GeneAll Biotechnology, Seoul, Korea) according to the manufacturer’s protocol. The DNA quality and quantity were examined using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Whole-genome sequencing was conducted with the Illumina NovaSeq 6000 platform with paired-end reads of 2 × 151 bp at LabGenomics Co. Ltd. (Seongnam, Korea). Phred scores of 20 or lower were removed from the total NGS paired-end reads with the CLC-quality trim tool included in the CLC Assembly Cell package (ver. 4.06 beta). The plastome was de novo assembled using two methods, GetOrganelle v1.7.6.1 and the modified dnaLCW method (Kim et al., 2015; Jin et al., 2020). Assembly errors and gaps in the assembled plastome were manually corrected by mapping raw reads with Minimap2 (Li, 2018). Gene annotation of the assembled plastome was performed using BLAST and Chloe v. 0.1.0 in GeSeq and manually curated using the Artemis annotation tool (Altschul et al., 1990; Carver et al., 2012; Tillich et al., 2017). A circular map of the S. prunifolia f. simpliciflora plastome was drawn using OGDRAW v 1.3.1 (Greiner et al., 2019).
The 45S nrDNA unit sequences of S. prunifolia f. simpliciflora were assembled using the modified dnaLCW method. The start and end positions for each 45S nrDNA subunit (18S, ITS1, 5.8S, ITS2, and 26S) were determined using BLAST. The 45S nrDNA sequence of Musa acuminate cv. Formosana (LC610757.1) was used as a reference. The 5S nrDNA sequence was assembled by mapping raw reads to the sequence of Arabidopsis thaliana (AF330993.1). After determining the 5S nrDNA sequence, the intergenic spacer (IGS) region was assembled by elongation, with this step repeated until the elongated sequence reached the next 5S nrDNA unit. The total 5S nrDNA unit with the corresponding IGS region was confirmed using BLAST (Altschul et al., 1990). In addition, a total of seven sequence-read-archive (SRA) data consisting of one Aruncus dioicus and six Spiraea (S. media, S. chamaedryfolia, S. × rosalba, S. alba var. latifolia, and two S. × billardii) were downloaded from NCBI GenBank with accession numbers ERR5555288, ERR5554804, ERR5554733, ERR5554594, ERR555281, ERR5554552, and ERR5554770, respectively, and used for assembly and annotation of the plastome and nrDNA transcription units via the assembly strategies described above.
Twenty-four plastome sequences in the tribe Spiraeeae and two plastome sequences of the genus Gillenia in the tribe Gillenieae (outgroup) were included in the phylogenetic analysis. A total of 76 protein-coding genes shared by 26 plastomes were used for the phylogenetic analysis. Each gene was aligned using MAFFT v. 7.427 with the --maxiterate 1000 option (Katoh and Standley, 2013), and then concatenated into a matrix. Phylogenetic analysis was performed using the RAxML v. 8.2.12 with 1,000 replicates and the GTRGAMMA model (Stamatakis, 2014). To compare the phylogenetic relationships of the genus Spiraea between plastome and nrDNA phylogenies, eight plastome and nrDNA sequences (seven Spiraea and one Aruncus) assembled in this study were used. The plastome sequences with only one copy of the IR region and entire 45S nrDNA sequences were aligned using MAFFT, after which RAxML was used for phylogenetic reconstruction. The options used for alignment and phylogenetic reconstruction steps were identical to those described above.
RESULTS AND DISCUSSION
A total of 22,962,984 reads were generated by means of whole-genome shotgun sequencing. Approximately 13.02% of the obtained reads were determined as the plastome reads with 2,643.92× coverage. The assembled plastome sequence of S. prunifolia f. simpliciflora was 155,984 bp in length with a GC content of 36.7% and with a quadripartite structure, consisting of 84,417 bp of a LSC region (GC content: 34.5%), 18,887 bp of a SSC region (GC content: 30.3%), and 26,340 bp of two IR regions (GC content: 42.5%). The plastome contained 113 genes, consisting of 79 protein-coding genes, 30 tRNA genes, and four rRNA genes (Fig. 1A). The assembled plastome sequence, BioProject, BioSample, and SRA data can be accessed via accession numbers OP874593, PRJNA904405, SAMN31842511, and SRR22385993, respectively. The assembled 45S nrDNA sequence was 5,848 bp in length with a GC content of 55.9%, consisting of 1,809 bp of 18S, 161 bp of 5.8S, and 3,397 bp of 26S which were separated by two ITS regions, 261 bp of ITS-1 and 220 bp of ITS-2 (Fig. 1B). The total length of the assembled 5S nrDNA unit was 512 bp, made up of 121 bp of a 5S transcription unit and 391 bp of an IGS region (Fig. 1C). The assembled 45S nrDNA and 5S nrDNA sequences can be accessed via accession numbers OP966298 and OP957414, respectively.
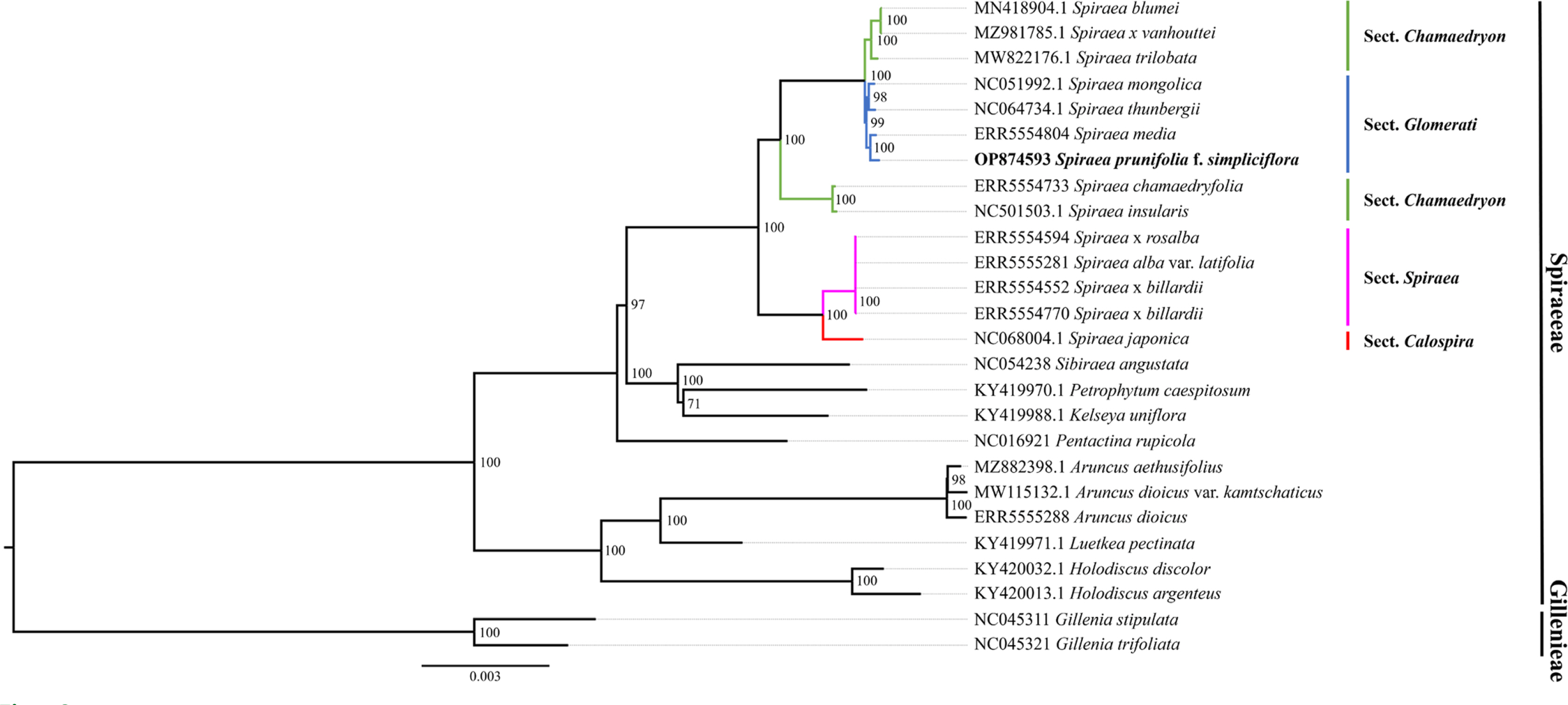
The phylogenetic result based on 26 plastome sequences indicated that the genus Spiraea was monophyletic (Fig. 2). In the tribe Spiraeeae, Sibiraea angustata, Petrophytum caespitosum, and Kelseya uniflora formed a single clade that was a sister to the genus Spiraea, and this result was consistent with previous studies (Suh et al., 2021; Park et al., 2022). Most clades in the genus Spiraea were well supported by nearly 100% bootstrap values, except for the sect. Spiraea. Sect. Glomerati, including S. pruniflora f. simpliciflora, S. thunbergii, and S. media, was nested within sect. Chamaedryon (Fig. 2); this phylogenetic relationship has previously been reported as well (Yu et al., 2018).
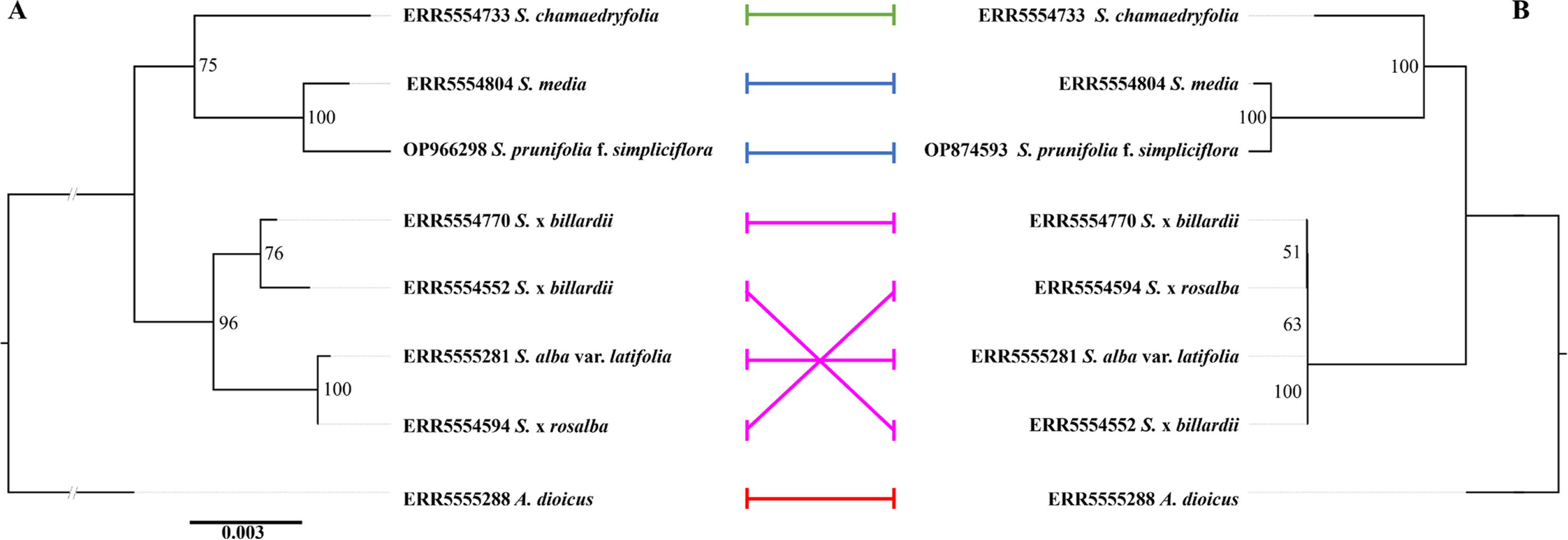
A comparison between the plastome and nrDNA phylogenies showed that S. prunifolia f. simpliciflora has a consistent position as a sister to S. media in both phylogenies (Fig. 3). However, the sect. Spiraea, composed of S. alba var. latifolia and its hybrid species, S. × rosalba and two S. × billardii, showed discordance between the two phylogenies owing to the different inheritance systems between the plastid and nuclear genomes (Zhang et al., 2012; Khan et al., 2014). In the phylogeny based on biparentally inherited nrDNA sequences, two S. × billardii formed their own subclade; S. alba var. latifolia formed a subclade with S. × rosalba (Fig. 3A), but they were not in the plastome-based phylogeny with relatively low branch support values (Fig. 3B). The plastome-based phylogeny showed a short branch length in the clade of S. alba var. latifolia and its hybrid species (Fig. 3B). The low resolution of plastome-based phylogeny may be caused by the maternal inheritance of the plastome in the genus Spiraea, providing some genetic evidence of the artificial hybridization of S. × rosalba and S. × billardii with S. alba as their maternal parent (Plants of the World Online, 2023). A comparison between plastome and nrDNA phylogenies may reveal the other inherited patterns of the two species originating from hybridization, S. × rosalba and S. × billardii. It is known that S. × rosalba was generated by hybridization between S. alba and S. salicifolia, and S. × billardii was generated from that between S. alba and S. douglasii (Plants of the World Online, 2023). The nrDNA phylogeny showed that two individuals of S. × billardii, whose paternal parent was S. douglasii, formed their own subclade, and S. × rosalba, whose paternal parent was S. salicifolia, formed a subclade with S. alba var. latifolia, suggesting that the nrDNA phylogeny provides the evolutionary patterns of the biparental inheritance, including the paternal inheritance.
Consequently, the infrageneric relationships of the genus Spiraea remain unclear (Oh et al., 2010; Lee and Hong, 2011), and conflicts between morphological and molecular evidence still exist (Yu et al., 2018). Further studies using both biparental nuclear and uniparental plastome data with extensive sampling will be needed to reveal the infrageneric relationships of the genus Spiraea. We believe that the complete plastome and nrDNA sequences of S. prunifolia f. simpliciflora will be useful in further studies to understand the phylogenetic relationships and the evolutionary history of the genus Spiraea, as well as the family Rosaceae.