The genus Adonis L. belongs to the family Ranunculaceae and is composed of perennial and annual herbaceous plants that are included in the tribe Adonideae under the subfam. Ranunculoideae (Nishikawa and Kadota, 2006; Ren et al., 2009). Approximately 26–30 species grow in the northern temperate zone including Asia, Europe, and North America, and some annual plants are known to be distributed from Southwest Asia to North Africa, as well as along the shores of the Mediterranean (Meusel et al., 1965; Cronquist, 1981; Mabberley, 1990).
After the initial attempt by Linnaeus (1753), De Candolle (1818) established the classification system, including the genus Adonis at the section level, in which 10 taxa of this genus were divided into the sect. Adonia DC., an annual species with 5–10 petals, 18–20 stamens, erect styles, and cylindrical or globular aggregated achenes, and the sect. Consiligo DC., a perennial species with 8–15 petals, 25–30 stamens, recurved styles, and globular aggregated achenes. There have been various arguments on the classification of the sect. Consiligo DC. For instance, unlike the sect. Adonia DC., this group was subdivided into subsection levels (Bobrov, 1937; Poschkurlat, 1977; Tamura, 1990, 1991) and some taxa were treated as an independent genus (gen. Adonanthe Spach) or section (sect. Ancistrocarpium Spach) (Spach, 1839).
Through a morphological study on 30 taxa of the genus Adonis L. distributed globally that represented a comprehensive study of this genus, Wang (1994a, 1994b) promoted sect. Adonia (annual plants) and sect. Consiligo DC. (perennial plants) to the subgenus Adonis (DC.) W. T. Wang and subgen. Adonanthe (Spach) W. T. Wang, respectively. Furthermore it was suggested that the subgen. Adonis should be further divided into 3 sections and 2 series, while the subgen. Adonanthe should be divided into 3 sections and 4 series, resulting in a new classification system with 1 genus, 2 subgenera, 6 sections, and 6 series.
The sect. Adonanthe W. T. Wang in the subgen. Adonanthe is divided into the series Amurenses Poschk., which is characterized by ovate, ovate deltoid, or elliptic shaped leaves and yellow or white petals; ser. Coeruleae Poschk. with oblong or ovate oblong shaped leaves and white or purple petals; ser. Apenninae Bobr. ex Poschk. with 3-pinnately compound leaves; and ser. Vernales Bobr. ex Poschk. with 3-pinnatisect palmately compound leaves. Son and Ko (2013) have supported this classification system within the sect. Adonanthe based on the morphology of aggregated achenes and the microstructure of the achene surface in the genus Adonis L. found in East Asia; however, most of the previous studies on the genus Adonis L. mentioned above focused on the classification system based on morphological differences, including those associated with leaf shape and achene characters, whereas a phylogenetic relationship-based classification system of the genus Adonis L. has not yet been established. In addition, Son (2015) indicated that, although leaf shape and leaf type in the genus Adonis L. could be used for identification in some species, they were highly variable among individuals and in most cases generally did not provide any information for phylogenetic analysis.
Because plants of the sect. Adonanthe have similar external morphological characteristics that are highly variable and objective taxonomic characteristics of the taxa within the genus have not been established, it is difficult to identify taxa and investigate their phylogenetic relationships. A. davidii Franch. is a Chinese endemic species described by Franchet (1885), but it was considered a synonym of A. brevistyla Franch. (Wang, 1980). Later, it was reestablished as an independent taxon by Wang (1994a), whereas A. brevistyla was treated as a synonym of A. davidii by Fu and Robinson (2001), which shows the different opinions about the taxonomic status of these taxa.
Adonis pseudoamurensis W. T. Wang (Korean name: Gaebok-su-cho) was introduced as a new species by Wang (1980) based on samples collected in Jilin, China (C. Chen 1204, PE), but it was later considered a synonym of A. ramosa Franch. (Korean name: Ga-ji-bok-su-cho) along with A. multiflora Nishikawa et Koji Ito (Korean name: Se-bok-su-cho) by the same author (Wang, 1994a). However, Nishikawa and Kadota (2006) regarded A. ramosa as a Japanese endemic species and Wang’s (1980) A. pseudoamurensis as a synonym of A. multiflora. In contrast, Lee et al. (2003) and Son and Ko (2012) recognized A. pseudoamurensis, A. multiflora, and A. ramosa as independent species. In addition, A. pseudoamurensis was treated as a variety of A. amurensis [var. pseudoamurensis (W. T. Wang) Y. N. Lee] (Lee, 2004, 2006). Thus diverse opinions have appeared about its taxonomic status.
Adonis coerulea Maxim. was described by Maximowicz (1877) based on samples collected from Gansu, China (N. M. Przewalski s.n., E). The species is characterized by plants glabrous; leaves oblong, 2–3 pinnately compound, with long petioles, densely arranged at the base of the stem; and petals purple. Based on the samples collected from Xizang, China (Z. Qing et al. 76-10534, PE), Wang (1980) designated the population with short pubescence on the stem and leaves, unlike glabrous f. coerulea, as f. puberula W. T. Wang, and later Wang (1994b) elevated it to var. puberula (W. T. Wang) W. T. Wang. However, Fu and Robinson (2001) considered var. puberula a synonym of the basic species and recorded it as a hairless plant except for pistils, adding further to the uncertainty of the taxonomic identity of the f. puberula population, which has been recognized for having short pubescence on the stem and leaves.
Recently, molecular data have been frequently used to establish a phylogenetic classification system. In particular, the ITS regions, which are nuclear DNA regions characterized by patterns of parental inheritance, evolve more rapidly than the coding regions, leading to higher levels of variation among closely related individuals. Thus, the ITS regions have been utilized to investigate inter-species and inter-genus relationships, as well as evolutionary trends and patterns in genetic variation (Baldwin, 1992; Álvarez and Wendel, 2003).
Previous studies on the genus Adonis L. in East Asia include sequence analyses of trnL-F and the ITS region sequenced from Japanese Adonis plants (Kaneko et al., 2008) as well as an ITS sequence analysis from Korean Adonis plants (Suh et al., 2002). Additionally, despite the taxonomic applications of various DNA sequence analyses, which have been completed by a number of taxonomists, the genus Adonis L. has only been used as a control for studying relationships with other taxa within the family (Hoot, 1995; Sun et al., 2001; Després et al., 2003; Cai et al., 2009). Furthermore, no representatives from the entire taxa of the sect. Adonanthe under gen. Adonis L. in East Asia have been phylogenetically classified.
Thus, in the present study, we investigated the phylogenetic relationships of the sect. Adonanthe of genus Adonis L. using sequences from the ITS region of nuclear ribosomal DNA and aimed to evaluate the validity of the intra-genus classification system more objectively. In addition, we also attempted to address the taxa with species delimitation problems.
Materials and Methods
Materials
Plant samples from 11 taxa with 21 representative accessions from the sect. Adonanthe of genus Adonis were either collected directly from the field by the participants in this study or obtained from specimens that were deposited in herbaria (HNHM, PE, and KUN) in Korea and China. Nuclear acid sequence information for A. vernalis L. and A. shikokuensis Nishikawa et Koji Ito were retrieved from GenBank. Data from GenBank were also added for the taxa with different taxonomic status among the sect. Adonanthe. A. annua L. belongs to the sect. Adonis of subgen. Adonis, and Trollius acaulis Lindl., T. asiaticus L., and T. chinensis Bunge were selected as the outgroup (Johansson, 1999). The collection data regarding the 13 taxa with 49 accessions and 4 taxa as the outgroup with 4 accessions that were used in the present study are summarized in Table 1.
DNA extraction, DNA amplification, and sequencing
Leaf samples collected from specimens or the field were stored in silica gel until completely dehydrated and crushed using a grinder (Mixer Mill MM200, Retsch, Haan, Germany) or a mortar, and total genomic DNA was extracted using a DNA extraction kit (MGTM Plant Genomic DNA Extraction SV mini, Macrogen, Seoul, Korea). Extracted DNA was assessed by electrophoresis using a 0.9% agarose gel. The reaction mixture utilized to amplify DNA using polymerase chain reaction (PCR) included 5 μL 10× Pfu DNA buffer, 1 μL 10 mM dNTP, 1 μL 10 pmol primers in each direction, 25–100 ng template DNA, 0.5 μL Pfu DNA polymerase, and 41.5 μL D.W. in a total volume of 50 μL. The primers used for DNA amplification were ITS4 and ITS5 (White et al., 1990). PCR conditions for gene amplification were as follows: the reaction began with a pre-denaturation step at 94°C for 5 min, followed by 35 thermal cycles of denaturation at 94°C for 30 sec, annealing at 52°C for 1.5 min, and extension at 72°C for 1.5 min, followed by a final extension at 72°C for 7 min. The PCR product was purified with a commercial kit (PCR Purification Kit, Macrogen, Korea), followed by cycle sequencing using a BigDye® terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA). Sequences were analyzed using the ABI PRISM 3730XL Analyzer (Applied Biosystems, USA). Sequences were deposited to GenBank and accession numbers were obtained (KU570388–KU570408).
Alignment of DNA sequences and analyses of phylogeny
DNA sequences for each sample in both the forward and reverse orientations were assembled and aligned using Geneious ver. 7.1.7 (Biomatters, Ltd., Auckland, New Zealand), and final alignments were manually verified. Maximum Parsimony analysis (MP) and Maximum Likelihood (ML) were conducted using PAUP* ver. 4.0b10 (Swofford, 2003). Heuristic search was carried out with ACCTRAN, MULPARS, and TBR branch swapping options turned on. To determine the support levels for the resulting cladograms, the bootstrap values (Felsentein, 1985) and posterior probabilities (PP) from the Bayesian inference were determined. Bootstrap values were obtained from 1,000 bootstrap replicates. To calculate the posterior probabilities using a Bayesian inference, a substitution model best fit to each set of sequence data was selected based on Akaike Information Criterion (AIC; Posada and Buckley, 2004) using Modeltest ver. 3.7 (Posada and Crandall, 1998). After the best model was selected, 2×106 generations were run with the Markov chain Monte Carlo method (MCMC; Hastings, 1970) using MrBayes ver. 3.1.2 (Huelsenbeck and Ronquist, 2001), and then posterior probabilities were calculated after discarding the burn-in sample (initial 500,000 generations).
Furthermore, a tree topology built using the Neighbor Joining method (NJ) (Saitou and Nei, 1987; Farris et al., 1995) and pairwise sequence divergence was calculated by the Kimura two-parameter method (Kimura, 1980).
Results
Sequences of nrDNA ITS region
The total DNA sequence length of the ITS region from 49 accessions representing 13 taxa in the sect. Adonanthe was 597–601 bp, and the total length of the aligned sequences was 604 bp. The length of ITS 1 and ITS 2 was 232–234 bp and 201–203 bp, respectively, showing that the length of ITS 1 was longer than that of ITS 2. The length of the 5.8S region was 164–165 bp, which was similar to that in other flowering plants (161–164 bp).
Pairwise distance between taxa calculated using Kimura’s two parameter method revealed that sequence divergence of the ITS regions across 53 samples, including the outgroup, was 0–9.873%; and sequence divergence among 49 samples, excluding the outgroup, was relatively high (0–6.106%). In the data set that included outgroups, taxa without intraspecific sequence variation included A. ramosa (RAM1, RAM3, RAM4, RAM5, RAM7, RAM8, and RAM11), A. amurensis (AMU6 and AMU7), A. shikokuensis (SHI1), A. pseudoamurensis (PSE4, PSE7, and PSE8), and A. multiflora (MUL4 and MUL5) (Table 1). The highest sequence divergence was observed between A. annua and A. amurensis (AMU1). Among the ingroups, A. davidii (DAV1) and A. amurensis (AMU3 and AMU5) were the taxa with the highest sequence divergence (6.106%).
Phylogenetic analyses
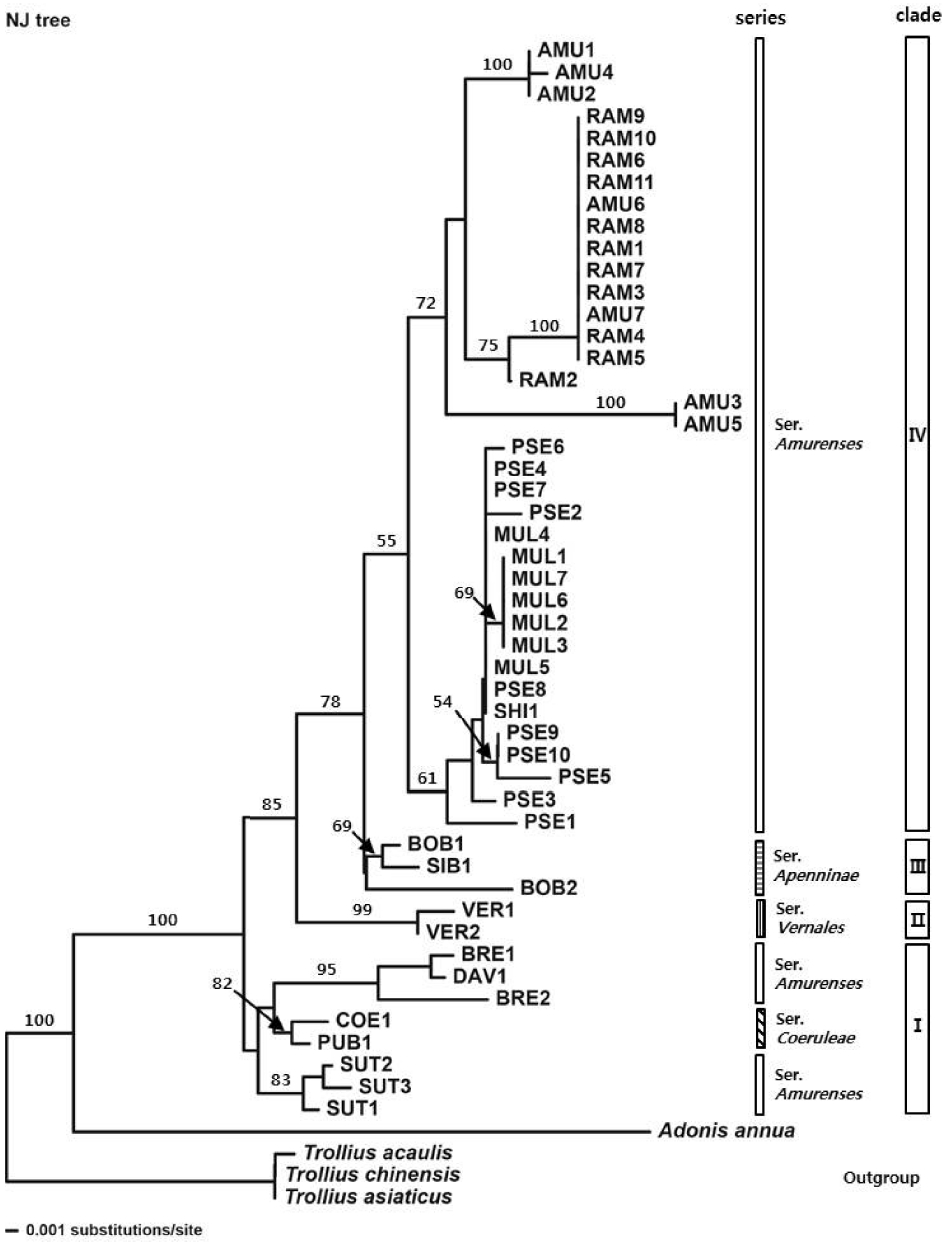
NJ analysis based on sequence divergence resolved the species of the Adonis sect. Adonanthe largely into four clades (Fig. 1). Clade I comprised A. sutchuenensis Franch., A. davidii, and A. brevistyla from ser. Amurenses and A. coerulea and A. coerulea var. puberula from ser. Coeruleae; it was resolved as the most basal, forming a sister group with other clades. Then A. vernalis from ser. Vernales (clade II) diverged, and clade III including A. bobroviana Simonovich and A. sibirica (Patrin ex DC.) Patrin ex Ledeb. from ser. Apenninae diverged after A. vernalis. Clade IV comprised two subclades, subclade 1 consisting of A. pseudoamurensis, A. multiflora, and A. shikokuensis, and subclade 2 including A. amurensis, and A. ramosa, which is congruent with the phylogenetic tree inferred from MP analysis (Fig. 2). Within the subclade 1, some accessions (PSE4, PSE7, PSE8, MUL4, and MUL5) possessed the same ITS sequences with A. shikokuensis (Table 1). Also, two accessions (AMU6 and AMU7) of A. amurensis were located in the same subclade as A. ramosa, showing a genetically close relationship between these taxa.
MP analysis of the ITS regions resulted in 34 equally parsimonious trees with a tree length of 170 steps, in which the CI (Consistency Index), RI (Retention Index), and RC (Rescaled Consistency Index) were 0.800, 0.931, and 0.744, respectively. The total length of the aligned sequences was 604 bp, of which, 115 were variable sites and 76 were parsimony informative sites.
In the parsimony tree (Fig. 2), the sect. Adonanthe of genus Adonis L. formed a monophyletic group consisting of 4 clades. The sect. Adonanthe had bootstrap values less than 50% for divergence of a clade (clade I) composed of A. sutchuenensis, A. coerulea, A. coerulea var. puberula, A. davidii, and A. brevistyla. In this clade, A. sutchuenensis formed a subclade with an 88% bootstrap value, and a sister subclade composed of A. davidii, A. brevistyla, A. coerulea, and A. coerulea var. puberula was formed. A. vernalis (clade II) diverged with a 100% bootstrap value, and a clade III composed of A. bobroviana and A. sibirica diverged with a bootstrap value less than 50%. In addition, a subclade composed of A. pseudoamurensis, A. multiflora, and A. shikokuensis and a subclade composed of A. amurensis and A. ramosa formed clade IV with a bootstrap value of 65%. Ser. Amurenses formed a polyphyletic group with representatives present in clade Iand clade IV.
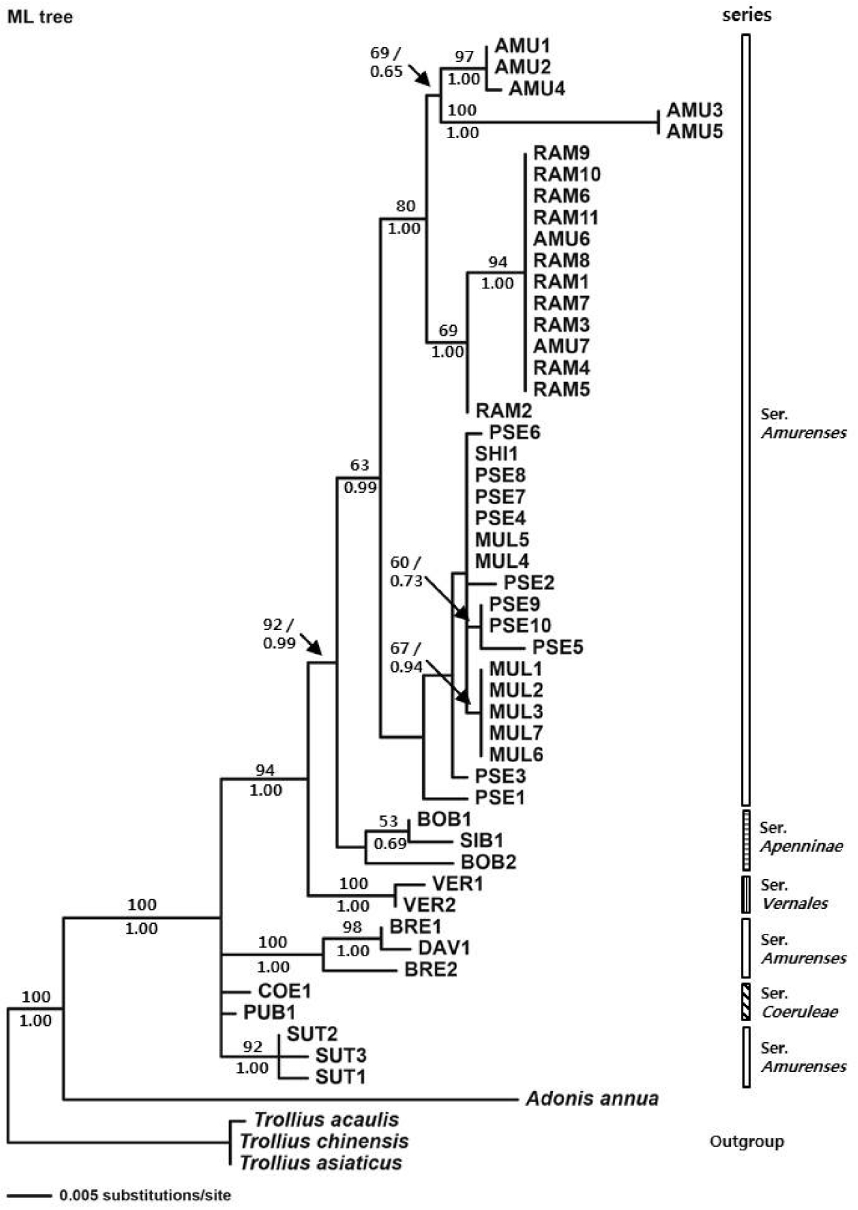
ML analysis (Fig. 3) showed monophyly of the genus Adonis sect. Adonanthe with bootstrap values of 100% and a Bayesian posterior probabilities value of 1.00. The clade I observed on the NJ and MP tree was not evident in the ML tree. A. vernalis diverged with a bootstrap value of 100% and a Bayesian posterior probabilities value of 1.00 (clade II). Clade III composed of A. bobroviana and A. sibirica formed a sister group with clade IV composed of A. pseudoamurensis, A. multiflora, A. shikokuensis, A. amurensis, and A. ramosa with a bootstrap value of 92% and a Bayesian posterior probabilities value of 0.99. A subclade composed of A. amurensis and A. ramosa within clade IV formed a sister group with a subclade composed of A. pseudoamurensis, A. multiflora, and A. shikokuensis with a bootstrap value of 63% and a Bayesian posterior probabilities value of 0.99.
Discussion
A phylogenetic study using ITS sequences was conducted for 13 taxa that belong to the sect. Adonanthe of genus Adonis provided valuable information for the classification of the taxonomically problematic section. Although the ITS trees mostly supported monophyly of ser. Vernales and ser. Apenninae, the taxa in ser. Amurenses did not form a monophyletic group (Figs. 1–3). The polyphyly of ser. Amurenses is probably due to genetic divergence caused by disjunctive distribution of the members in the series (A. davidii, A. brevistyla, and A. sutchuenensis in mid-western China and other taxa in northeastern China, Korea, and Japan). These results are inconsistent with the classification system proposed by Wang (1994a, 1994b).
Meanwhile, the taxa in the clade I (Figs. 1, 2), which is composed of A. davidii, A. brevistyla, and A. sutchuenensis in ser. Amurenses and A. coerulea and A. coerulea var. puberula in ser. Coeruleae, grow in fruticeta or grasslands at an elevation of 1,500–3,000 m in the mid-southern, southwestern, and western areas in China and are characterized by elongated and lignified rhizomes. This is in contrast with those taxa found in northeastern China, Korea, and Japan, including the distinct clade IV (A. amurensis, A. ramosa, A. pseudoamurensis, A. multiflora, and A. shikokuensis), which have short rhizomes and are surrounded by fibrous roots. In the strict consensus tree (Fig. 2), however, clade I composed of A. sutchuenensis, A. coerulea, A. coerulea var. puberula, A. davidii, and A. brevistyla collapsed, which is consistent with the results from the ML analysis (Fig. 3). Despite forming a clade in the NJ analysis (Fig. 1), either the bootstrap value supporting the clade I was low or the taxa examined did not form a monophyletic group depending on the analysis method implemented. Thus, further studies are needed to resolve the phylogenetic relationships among these groups in relation to the evolutionary trends of morphological characters.
There have been many different opinions regarding the taxonomic identities and phylogenetic relationships of the taxa that belong to the sect. Adonanthe. It is known that the members of sect. Adonanthe have similar morphological characters with severe variations, which hindered establishment of dependable taxonomic characters within the section. A. davidii and A. brevistyla formed a subclade with bootstrap value of 99% in the MP analysis (Fig. 2), which revealed that they are genetically related taxa. It is noticeable that BRE1 and BRE2 in A. brevistyla did not form a clade in all ITS trees (Figs. 1–3). Depending on the taxonomists, A. davidii and A. brevistyla have been considered either conspecific (Wang, 1980; Fu, 2000; Fu and Robinson, 2001) or treated as independent species (Wang, 1994a). Since it is possible that A. davidii was misidentified as BRE1 in the present study, a taxonomic study based on the original description and a type specimen should be performed to validate the identities of these taxa. Meanwhile, Wang (1994a, 1994b) suggested that A. davidii was the most primitive species in the sect. Adonanthe. However, clade I collapsed in the strict consensus tree, making the group consisting of A. davidii, A. sutchuenensis, A. coerulea, and A. coerulea var. puberulea is paraphyletic, which is inconsistent with the taxonomic opinion of Wang (1994a, 1994b). Nevertheless, A. davidii is positioned at the base of sect. Adonanthe in MP, NJ, and ML analyses (Figs. 1–3). In addition, the species has ancestral characters, including long rhizomes, long petioles, oval lobules, and short persistent styles with recurved ends and is found in the southwestern region of China. This region is the center of origin for the genus Adonis L., and for this reason it has been considered to be the progenitor species of the sect. Adonanthe (Wang, 1994a, 1994b; Son, 2015).
Adonis coerulea and A. coerulea var. puberula were clustered together in the NJ analysis (Fig. 1), yet they did not form a clade in MP and ML trees (Figs. 2, 3). In some cases, A. coerulea var. puberula has been considered as a synonym of A. coerulea by Fu and Robinson (2001); thus, use of multiple samples collected from diverse areas and employment of phylogenetically useful DNA regions is imperative in order to confirm their taxonomic identities.
Although A. sutchuenensis, an endemic Chinese species, is superficially similar to A. amurensis, it is clearly distinguished by following morphological characters: the leaf shape is ovate pentagon and the sepal is shorter and narrower than the petal (Franchet, 1894). Within the ITS trees, multiple accessions representing A. sutchuenensis formed a strong monophyletic group, supporting the species is clearly distinct from other taxa within the sect. Adonanthe; therefore, it is reasonable to treat it as an independent species.
Adonis vernalis formed a sister clade with the group composed of clade IV (including representatives in ser. Amurenses found in northeastern China, Korea, and Japan) and clade III (ser. Apenninae distributed in mid-western China, Mongolia, Russia, and Kazakhstan), indicating that it is directly related to these groups (Figs. 1–3). In the sect. Adonanthe, A. vernalis is the only taxon distributed in Europe and Central Asia; therefore, it is thought that A. vernalis diverged and formed a distinct species when it moved from the western Himalayas to Europe. This divergence within the genus Adonis L. appears to support the views that the plants of the sect. Adonanthe (excluding A. vernalis) underwent speciation either after being transmitted along the route from southwestern China to Eastern Siberia via the northeastern Chinese region or from southwestern China to North Asia (Wang, 1994a, 1994b; Son, 2015).
Although the support for the clade is not sufficient, all ITS trees recognized monophyly of ser. Apenninae, which is supported by distinctive morphological characters such as lack of petioles (all other taxa of the sect. Adonanthe have petioles). Phylogenetic analysis, however, revealed that A. bobroviana is not monophyletic. The morphological characteristics of A. bobroviana, including glandular hairs on the stem and leaf, persistent style characterized by being recurved in a hook shape, and parallel achene surfaces, are clearly distinct from those of A. sibirica (Fu and Robinson, 2001; Son and Ko, 2013).
Adonis ramosa was treated either as a Japanese endemic species (Nishikawa and Kadota, 2006) or the same species as A. pseudoamurensis, which is found in Korea and China (Wang and Liu, 1988; Wang, 1994a, 1998; Fu and Robinson, 2001; Park, 2007). Additionally, A. pseudoamurensis was considered to be the same species as A. multiflora found in Korea and Japan (also reported in China; Nishikawa and Kadota, 2006); thus, there has been confusion concerning taxonomic identities of the taxa. The MP, NJ, and ML analyses of the ITS sequences distinguished A. ramosa from A. pseudoamurensis and revealed that A. multiflora was a distinct clade (Figs. 1–3). Morphologically, A. ramosa has hairs on the abaxial side of the leaf and its sepal length is similar to that of the petals. In contrast, A. pseudoamurensis and A. multiflora have no hair on the abaxial side of the leaves and the sepals are shorter than the petals. In addition, since A. pseudoamurensis has distinctive traits from A. multiflora with respect to the morphology of the leaf apex and lobule and microstructure of the achene surface (Son and Ko, 2012, 2013), it is reasonable to treat these species as independent taxa. With respect to chromosome number in the somatic cells, A. ramosa is a tetraploid plant with 2n = 32 (Nishikawa, 1988, 1989), whereas A. pseudoamurensis is a diploid plant with 2n = 16 (Son, 2011). Thus, it is unlikely that A. ramosa is distributed in Korea and China (Wang and Liu, 1988; Wang, 1994a, 1998; Fu and Robinson, 2001; Park, 2007). Instead, it appears to be reasonable to consider the species to be a Japanese endemic species (Suh et al., 2002; Lee et al., 2003; Nishikawa and Kadota, 2006). On the other hand, A. ramosa and A. amurensis formed a subclade with a bootstrap value of 75% in the MP tree (Fig. 2), indicating they are closely related taxa. However, the Korean A. amurensis and Japanese A. amurensis groups formed distinctive subclades. Interestingly, the Japanese A. amurensis accessions and the accessions of A. ramosa (RAM1, RAM3, RAM4, RAM5, RAM7, RAM8, and RAM11) had identical DNA sequences (Table 1). Geographically, A. ramosa is distributed in Hokkaido and northern to mid Honshu (Nishikawa and Kadota, 2006), and this area overlaps with that of A. amurensis in the eastern Hokkaido region (Nishikawa and Takeshima, 1992; Nishikawa, 1994). This is consistent with the theory of Kaneko et al. (2008), which speculated that A. ramosa should be an autotetraploid and the diploid progenitor of A. ramosa should be A. amurensis from the Hokkaido region. Since only 2 ITS ribotypes in A. ramosa were examined (Table 1), the data may not be sufficient to determine the process of speciation. However, the geographical distribution of organisms related to their ITS ribotypes was clearly distinct (Kaneko et al., 2008), implying the identical DNA sequence between the Japanese A. amurensis and A. ramosa appears to be due to a relatively recent speciation event.
For the Korean A. amurensis, the MP, NJ, and ML analyses divided the group into two subgroups (Figs. 1–3), which was attributed to the differences in the insertion/deletion of sequences (Table 1). However, no clear relationship was observed between the geographical distributions of these individuals and the ribotypes of each sample.
The ITS sequence analysis of A. pseudoamurensis revealed high levels of variation within the species (Table 1). This result is consistent with the report of Suh et al. (2002) that A. pseudoamurensis, unlike A. amurensis and A. multiflora, showed a high level of genetic variation in RAPD analysis. The high genetic variation in A. pseudoamurensis is probably the result of adaptation to diverse habitats in wide geographical areas of the species which ranges from northeastern China to southern part of the Korean Peninsula including islands on the west to south coast as well as lowland regions. Meanwhile, ITS trees indicated that A. pseudoamurensis is clearly distinct from A. amurensis (Figs. 1–3). In morphology, A. amurensis has an unbranched stem and its leaves emerge after the inflorescence has developed, whereas A. pseudoamurensis has a branched stem and its leaves and flowers develop at the same time; these differences together with phylogenetic data further validate recognition of A. pseudoamurensis as an independent species (Lee et al., 2003; Son and Ko, 2011, 2012) rather than as a variety of A. amurensis (Lee, 2004, 2006).
Some accessions of A. multiflora formed a subclade with a bootstrap value of 63% in MP analysis (Fig. 2). Additionally, the Sangumburi (MUL4) and Eorimok (MUL5) accessions were similar to A. shikokuensis that is distributed in Honshu, Shikoku, and Kyushu in Japan as well as to other groups (PSE4, PSE7, and PSE8) of the Korean A. pseudoamurensis (Figs. 2, 3, Table 1). This result suggest that A. shikokuensis might be related with either A. pseudoamurensis or A. multiflora, contradicting the hypothesis proposed by Nishikawa and Ito (2001), who suggested A. shikokuensis diverged from A. ramosa. However, further studies including more diverse samples and additional molecular markers are necessary to resolve the relationship among these taxa and obtain a reliable phylogenetic tree.