INTRODUCTION
Within the Asteraceae family, genus Chrysanthemum L., a well-known genus possessing various cultivars, is distributed primarily in Eastern Asia and the temperate climate zone of Siberia (Kadereit and Jeffrey, 2007; Plants of the World Online, 2019). About the circumscription of the species of the genus Chrysanthemum, many researchers have traditionally argued and proposed several classification systems to identify regional wild Chrysanthemum (Makino, 1903; Hu, 1958; Kitamura, 1978; Anderson, 1987; Trehane, 1995; Kim et al., 2003). Korean Chrysanthemum, is generally classified into three species of Chrysanthemum zawadskii complex, Chrysanthemum indicum L., and Chrysanthemum boreale Makino. The former C. zawadskii have ray flowers of white or pale pink, unlike the latter two species which have yellow ray flowers (Lee, 2003, 2006; Kim et al., 2014). However, the genus Chrysanthemum has a wide range of morphological variations, particularly in the size, shape, and anatomical structure of leaves, and these variations result in the ambiguity of the species circumscription of the genus (Oh et al., 1994; Kim and Tobe, 2009; Kim et al., 2014; Kang and Kim, 2020).
The basic chromosome number of the genus Chrysanthemum is x = 9, which indicates a wide range of ploidy (Liu et al., 2012). To date, a series of polyploids: diploids (2n = 2x = 18), tetraploids (2n = 4x = 36), pentaploids (2n = 5x = 45), hexaploids (2n = 6x = 54), and decaploids (2n = 10x = 90), have been reported in the Korean Chrysanthemum (Lee, 1967, 1969; Kim et al., 2003, 2008). In case of Chrysanthemum coreanum (H. Lév. & Vaniot) Nakai, a member of the C. zawadskii complex restricted to Mt. Hallasan on Jejudo Island, has been confirmed to be a decaploid (10x = 90) and not an octoploid (2n = 8x = 72) (Kim et al., 2004). Because ploidy has regarded as a vital factor related to plant evolution (Leitch and Bennett, 1997; Te Beest et al., 2012), the discovery of various ploidy populations has important implications for the study of the genus Chrysanthemum. Especially, the presence or absence of diploid populations can play a role in resolving plant evolution via polyploidy. The precise confirmation of ploidy levels at the population scale is crucial to accurately understanding the speciation and evolutionary history of the genus Chrysanthemum.
For a very long time, chromosome numbers have been used as a cytogenetic characteristic to explain the most basic genetic properties of living organisms (Mayrose and Lysak, 2021), and karyotypes include the number, shape, and size of chromosomes, have been generally used as one of important tools for species identification (Heslop-Harrison and Schwarzacher, 2011). The chromosome-based researches have been conducted on various species of the genus Chrysanthemum, especially the Japanese, Chinese, and Taiwanese species (Tanaka and Shimotomai, 1961; Wang et al., 1993; Kondo et al., 1999; Kim et al., 2004), and has provided us with cytogenetic insights to understand the complex relationships among the species and different ploids.
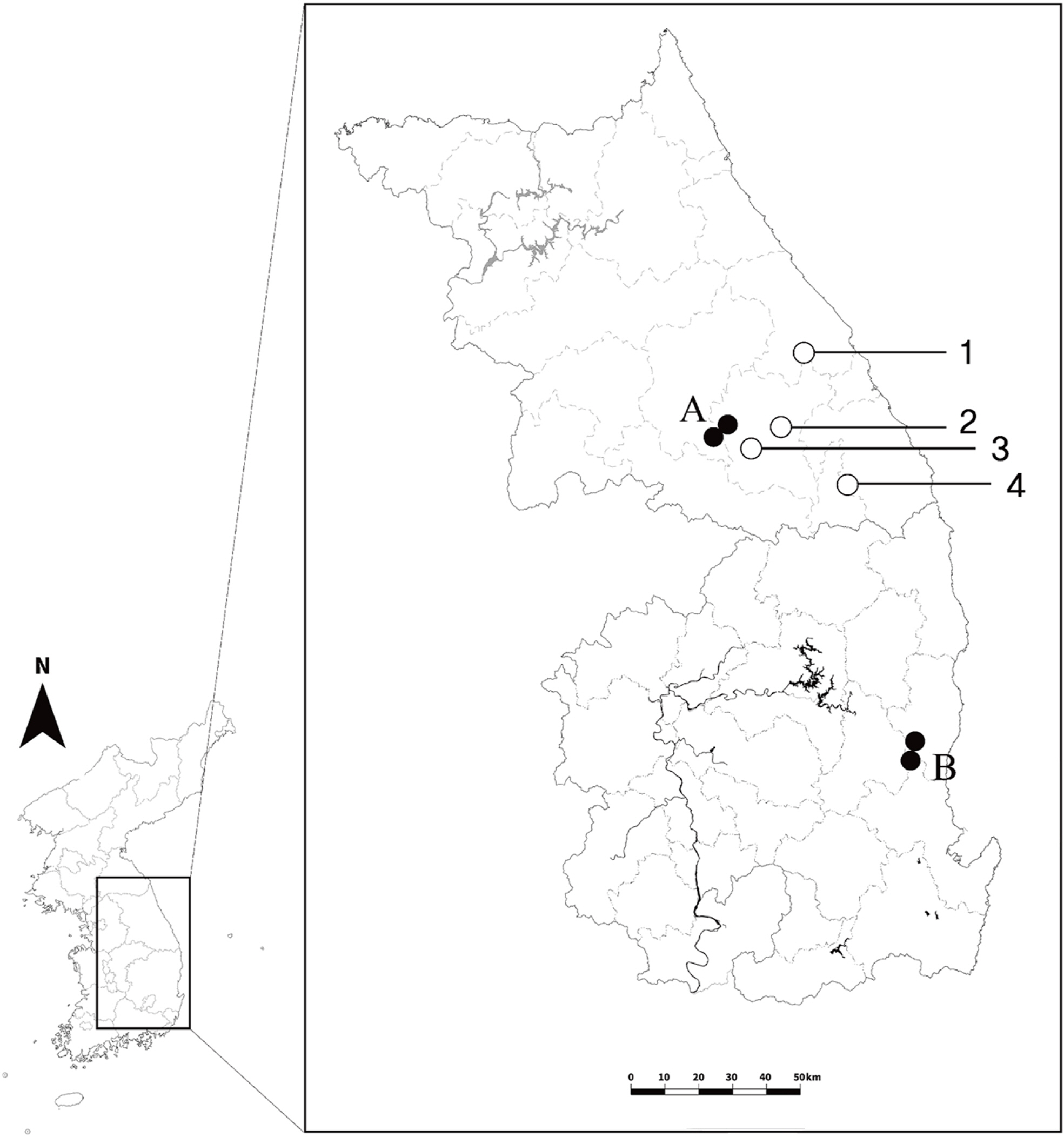
Chrysanthemum zawadskii Herbich is the only widespread species in the genus reported from Europe to Asia, and it forms a species complex of different ploidy levels and various infra-specific taxa in Korea although its correct taxonomical status is still being debated. Considering the first discoveries in the diploid populations of this species (sites A and B) (Fig. 1) (Kim et al., 2003), the successive ploids from diploid to decaploid were completed, and increasing the probability of understanding the evolutionary process in the genus. However, the number of diploid populations is still restricted than that of the tetra- and hexaploid populations which are widely distributed across Korea. Therefore, it still makes a limitation for the discussion on the diversity of the C. zawadskii complex as well as the genus Chrysanthemum.
During our recent investigation, we identified new populations of diploid C. zawadskii native to Korea. In this study, we describe the morphological and cytological characteristics of these new populations along with their relationships.
MATERIALS AND METHODS
Plant materials and morphology observation
Four wild populations of C. zawadskii were identified by morphological characteristics as described by Lee (2003, 2006) and were collected from the top or ridge of a mountain before flowering season (except for one population; Miin Falls was collected after flowering) in 2022 in Korea (Figs. 1, 2, Table 1). All the collected materials were transplanted and nurtured as living samples in a greenhouse at Chungbuk National University (Cheongju-si, Chungbuk, Korea) to confirm their morphological and chromosomal characteristics. A total of eight characteristics, including head flower size, color, and leaf size were investigated and measured from more than three individuals per population based on their flowering conditions to compare the morphology among the populations (Table 2).
Fixation of the root tip
According to Kim et al. (2003), healthy root tips were cut approximately 2–3 cm from the living material. The root tips were soaked in 10 mL 0.002 M 8-hydroxyquinoline solution for 2 h at room temperature (RT) for the pretreatment, and then it was carried out in a fixative solution (99% acetic acid:99.9% ethanol = 1:3 v/v) for 30 min on ice. The fixed roots were stored in 70% ethanol (99.9% ethanol:distilled water = 7:3 v/v) at –20°C until chromosome observation.
Chromosome observations and karyotype analysis
According to the protocols of Fan (1965) and Kim et al. (2003), the fixed root tip was dissociated for 90 s at 65°C in 1N HCl and placed on a slide glass. After being crushed, the root tip tissue was then separated and stained using 1% acetoorcein for 35 min at RT in a humid chamber. The mitotic metaphase chromosomes were observed under an optical microscope (Olympus BX50; Tokyo, Japan). To get the representative karyotype of each population, total of four individuals were randomly selected from new diploid populations and at least three slides per individual were made for observation. Finally, over three cells in the mitotic metaphase stage were analyzed for karyotyping per slide.
For karyotype analyses, high-quality magnified images were captured and edited using Adobe Photoshop CC (Adobe Systems, San Jose, CA, USA). The chromosome length was measured using KaryoMeasure software (Mahmoudi and Mirzaghaderi, 2023), and the karyotype was analyzed as per the standards set suggested by Levan et al. (1964). Based on the ratio of short arm to long arm and the centromere locations of the chromosome, the karyotypes were identified and described as follows: m, metacentric chromosomes; sm, submetacentric chromosomes; st, sub-telocentric chromosomes; t, telocentric chromosomes. In case of the chromosome with satellite, it was added an asterisk in the figure and table.
Flow cytometry analysis
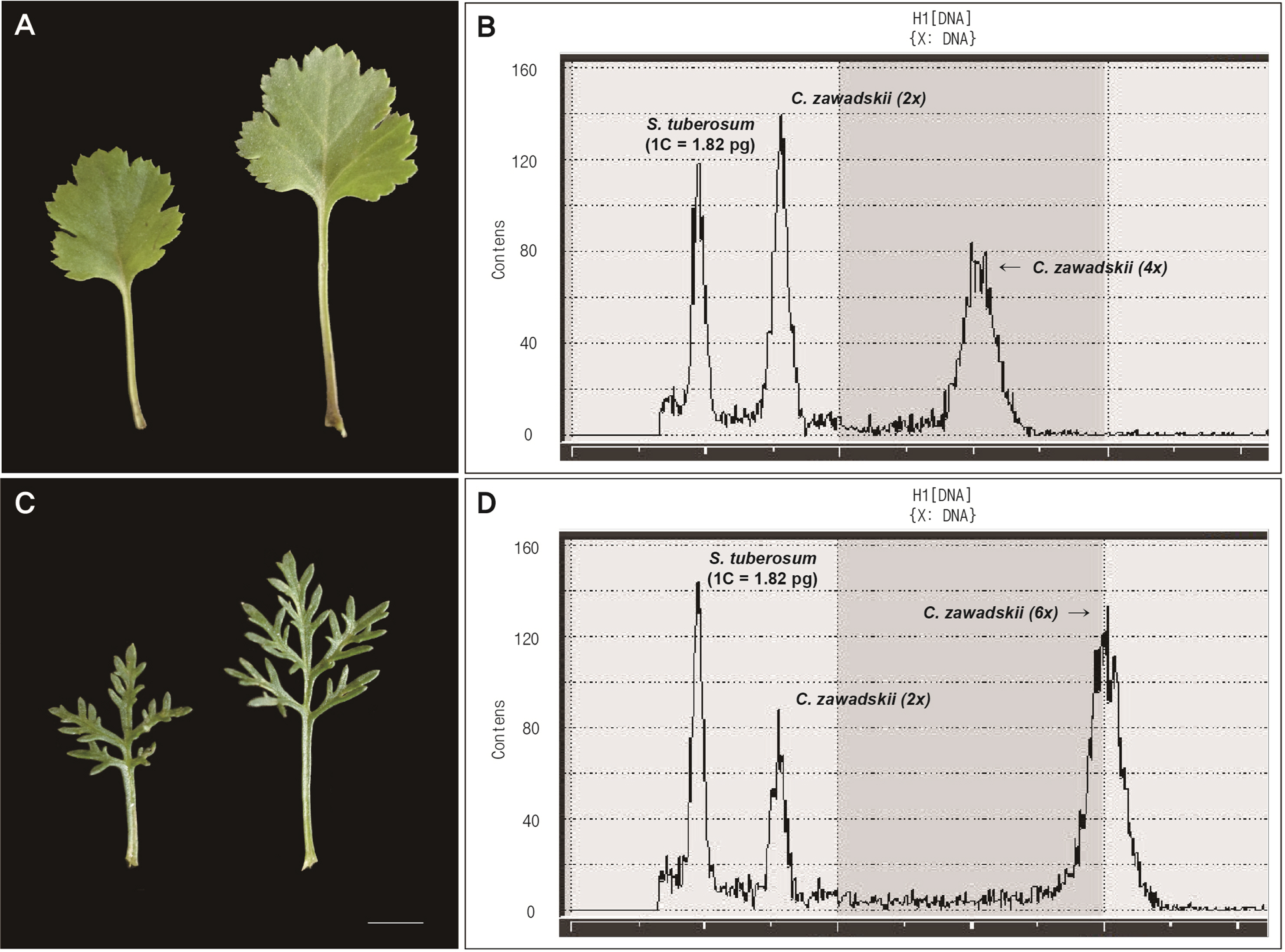
The ploidy levels of the diploid Chrysanthemum populations were measured with a tetraploid Solanum tuberosum (1C = 1.82 pg) (Valkonen et al., 1994) as a standard reference. In a Petri dish placed in ice, leaves of Chrysanthemum individuals and standard reference were chopped with a sharp blade using 500 μL of nuclei extraction buffer (CyStain UV Precise P kit, Sysmex Partec, Munster, Germany). Only the extract was filtered with non-sterile CellTrics Filter Green 30 μm (Sysmex Partec), the nuclei were stained with 2,000 μL staining buffer from the CyStain UV Precise P kit (Sysmex Partec). Ploidy level was measured at least three times per sample and recorded from each sample using CyFlow Cube 6 (Sysmex Partec). In case of the sympatric population Mt. Seokbyengsan, additionally found tetraploid and hexaploid were also measured together with standard reference and diploid populations for genome size estimation.
RESULTS
Morphological characteristics of diploid C. zawadskii
The morphological features of the four newly discovered diploid populations of C. zawadskii were compared even though the flower characters of Miin Falls population were not available due to late collection after flowering season. The results confirmed the existence of variations in flower size, color, and leaf shape (Fig. 3, Table 2). The flower heads were light purple-to-white in color. The flowers collected from Mt. Seokbyeongsan were very light purple until the end of the flowering season, whereas Mt. Gakhuisan and Mt. Baekisan populations had mostly white flowers. The head flower size of plants from the population of Mt. Gakhuisan was bigger than that of the other populations. Although the leaf length and width varied among the four populations, the length/width ratios ranged between 0.9 to 1.1.
Counting chromosome numbers and karyotype analysis
Based on the observations of mitotic metaphase chromosomes, it was confirmed that the four newly found populations were diploid (2n = 2x = 18) (Fig. 4) without exception. The representative karyotype of each population was confirmed (Fig. 5, Table 3) and grouped into three types: (1) 2n = 11m + 5sm + 2st* found on Mt. Seokbyeongsan (pop. 1) and Mt. Baekisan (pop. 3); (2) 2n = 12m + 4sm + 1sm* + 1st* found on Mt. Gakhuisan (pop. 2), and (3) 2n = 10m + 6sm + 2st* found on Miin Falls (pop. 4). Although the representative karyotypes of pop. 1 and 3 were same, the chromosome shape of 9th pair was different as 1 m + 1 sm and 1 sm + 1 st, respectively. A pair of secondary constrictions were identified in all individuals from different groups investigated in the current study.
Genome size estimation of the sympatric population
In Mt. Seokbyeongsan (pop. 1), we additionally found the tetraploid (2n = 4x = 36) and hexaploid (2n = 6x = 54) in the same place with a few meters apart from each other. Although it was difficult to clearly distinguish the diploid and tetraploid by morphological differences (Figs. 2a, 6), the hexaploid was clearly recognized by deeply split lobes. And it was confirmed that the chromosome number and genome size are increased according to its ploidy level (Table 4).
DISCUSSION
The Chrysanthemum zawadskii complex has been reported in Korea, with ploidy ranging from diploid (2n = 2x = 18) to decaploid (2n = 10x = 90) (Lee, 1967, 1969, 1975; Kim et al. 2004). Diploid can play a role in understanding evolutionary processes, especially in plant groups that underwent polyploidization during speciation. To date, four diploid C. zawadskii populations have been reported from two regions in Korea (sites A and B) (Fig. 1) (Kim et al., 2003), in contrast to polyploids that are widely spread over the Korean Peninsula. This was, however, insufficient to explain the evolutionary history of this highly diversified plant group. The current study further identified four new diploid populations of C. zawadskii from natural habitats in Korea (Figs. 2, 4). From the result, it could understand the variation of the Korean C. zawadskii complex in the diploid level together with the already available information of previously reported population. The habitat of some of these new populations was located on mountain peaks, valleys, or ridges of approximately 1,000 m, and they are all included in the Taebaek Mountain Range that runs along the eastern coast of Korea: (1) the diploid population of Mt. Seokbyeongsan was found in a rocky area at the top of the mountain, (2) The population of Mt. Gakhuisan was found in the cracks of the rocks on the cliffs, (3) The population of Mt. Baekisan, a limestone-rich mountain, was sympatric with Chrysanthemum boreale Makino (2n = 18), and (4) The population of Miin Falls was found on the cliffs next to the waterfall in the vicinity of the canyon. These populations are also found in limestonerich areas (Kwon et al., 2002; Song et al., 2016; Kim et al., 2021). The distances between the populations ranged from 24 to 77 km, and the populations of Mt. Gakhuisan and Mt. Baekisan were the closest to each other. These were close to the previously reported site A (Gwangha-ri of Jeongseon-gun and Baekun-ri of Pyeongchang-gun, Gangweon-do, Korea) but distinct from site B (Dalsan and Okgye of Yeoungdeok-gun, Gyeongsangbuk-do, Korea) (Fig. 1). In particular, (1) Mt. Seokbyeongsan live together with tetraploids (2n = 4x = 36) and hexaploids (2n = 6x = 54) that is the first report from this study. Each population lived a few meters apart from each other. However, it is difficult to identify the morphological differences between the diploid and tetraploid population (Fig. 6, Table 4). Hexaploid was relatively easily recognized from them by its leaves with finely lobes.
It was revealed that the head flower size of the newly discovered diploid populations ranged from 2.89 to 3.50 cm. This is smaller than previously reported populations of the Korean native C. zawadskii, even though their ploidy information was not provided (Kim et al., 2014). The new diploid populations were actually collected before flowering season (except for Miin Falls population), and then transplanted into a greenhouse without ploidy information at first. After stabilizing in the greenhouse condition, it was confirmed the ploidy level using the number of somatic chromosomes. Using these confirmed plant samples, the morphological features were investigated during the flowering season of the same year. Considering the morphological variations in the genus Chrysanthemum, it could suspect the probability of morphological differences between native and greenhouse conditions, but it was not critical limitation to compare the morphological characters among these new diploid populations because they were grown up in the same condition.
Karyotypes of diploid individuals of Chrysanthemum are known to be very diverse even within the same population (Kim et al., 2003). All of the representative karyotype of each population found in this study was included in the previously reported types, however, the number and location of secondary constrictions were differed. It has been generally regarded that structural changes on chromosomes constitute a vital evolutionary mechanism contributing to diversification and speciation, and are especially associated with hybridization and polyploidization in angiosperms (Dowrick, 1952; Rana, 1965; Lee, 1975; Kim et al., 2003; Weiss-Schneeweiss and Schneeweiss, 2012; Kang and Kim, 2020). It is necessary to determine how these diverse karyotypes exist and evolve within the same population and species. It is expected that the discovery of multiple populations of diploid C. zawadskii will provide evidence and suggestion to resolve this existing issue.
From the present study, we found new diploid populations of Chrysanthemum zawadskii from the natural habitats of Korea and described the morphological and cytological characteristic. They were all diploids with chromosome number 2n = 18 and diverse karyotypes among the populations. Among them, the sympatric population, where three different ploidy levels are distributed together, was also found at first. It is expected that this finding will contribute to understanding this extensively diverse genera which have experienced the polyploidization and hybridization during their evolution.