Polyploidy has long been recognized as an important mechanism of the plant evolution (Grant, 1981; Soltis et al., 1993; Thompson et al., 1997; Ramsey and Schemske, 1998; Peirson et al., 2012), driving genomic novelty and acting as one of the primary mechanism of sympatric speciation (Otto and Whitton, 2000; Wood et al., 2009). Besides harmful effects of polyploidy on fertility and fitness due to mitotic and meiotic abnormalities, and genomic instability (Comai, 2005), polyploidy has also adaptive potential allowing some new polyploids to survive and evolutionary diverge (Soltis et al., 2009; te Beest et al., 2012). Genome duplication is often accompanied by a creation of reproductive barriers through multiple mechanisms including a transition from crosspollination to self-pollination or even from sexual to asexual reproduction, shifts in phenology, and spatial segregation (Comai, 2005; Kolář et al., 2017; van der Peer et al., 2017).
Increased genome size and genetic variation can also potentially affect the morphology and physiology of newly formed polyploids, and these changes might lead to niche differentiation (Ramsey, 2011; te Beest et al., 2012; Madlung, 2013; Sonnleitner et al., 2016). Adaptive potential of polyploids is, however, expected to depend on the route of polyploid origin. Polyploids are traditionally classified into the two major types, i.e., autopolyploids and allopolyploids (Kihara and Ono, 1926). It has been supposed that allopolyploidy that include two steps, hybridization and genome duplication, provide the polyploid an initial selective advantage because hybridizing species are already diverged (Comai, 2005). However, polyploidization both in allo- and autopolyploids can also lead to changes in gene expression and consequently to changes in ecology and it seems presently that these changes are rather species-dependent (van der Peer et al., 2017).
Owing to increased genetic variation in polyploids, it has been repeatedly proposed and experimentally proven that polyploid might tolerate newly created, disturbed or harsh environments (Ramsey, 2011; Manzaneda et al., 2012; Hao et al., 2013; Madlung, 2013). Indeed, it has been evidenced many times that polyploids are ecologically distinct from their diploid relatives and often occupy at least partially separate geographic ranges (e.g., Müntzing, 1936; Grant, 1981; Xie-Kui et al., 2008; Šafářová et al., 2011). However, several recent papers on large data sets questioned evidences for the supposed broader ecological tolerances or increased tolerance of polyploids to harsher environments because in most studied examples high niche overlap or absence of niche shifts between diploids and polyploids were found (Glennon et al., 2014; Marchant et al., 2016). It is clear that whether polyploidy promotes niche differentiation require further research (Parisod and Broennimann, 2016).
Allium L. is one of the largest genera of monocots (Friesen et al., 2006; Li et al., 2010). Currently, the number of species within the genus is estimated at 920 (Seregin et al., 2015). Mixed-ploidy species—taxa containing at least two cytotypes—represent 16% of all plant species based on chromosomal surveys (Rice et al., 2015) while their proportion comprise ca. 19% in the genus Allium (Hanelt et al., 1992). Understudied cytotype variation can lead to the underestimation of species richness (Soltis et al., 2010) and incorrect estimation of niche breath and geographic distribution (Wefferling et al., 2017). A better understanding of polyploid complexes or closely related taxa differing in ploidy level thus require basic cytogeographic data that help together with morphological data to resolve taxonomic issues and even enhance genetic approaches that may be confounded by cryptic and frequently also reticulate speciation (Soltis et al., 2007; Suda et al., 2007; Grusz et al., 2009).
One example of closely related taxa differing in ploidy level with hitherto unsolved taxonomy and limited knowledge of cytotype composition and distribution pattern are Allium thunbergii G. Don and A. sacculiferum Maxim., belonging to section Sacculiferum P. P. Gritz., of subgenus Cepa (Mill.) Radic (Friesen et al., 2006; Li et al., 2010) that occur throughout Eastern Russia, Eastern China, Korea, and Japan (Choi and Oh, 2011). Although both species do not have any information about ploidy levels by the original authors, A. thunbergii, originally described from Japan, was reported its chromosome numbers to 2n = 16 (2x), 32 (4x), or 48 (6x) in Japan (Kurita, 1952; Noda and Watanabe, 1967; Hoshiya, 1982; Hotta, 1998), while A. sacculiferum was described from Russia (Far East) and known as a tetraploid species of 2n = 32 in Russia (Probatova et al., 2013). It is still unclear if these closely related taxa could be considered as two separate species, especially there has been much confusion between A. thunbergii var. thunbergii and A. sacculiferum (Choi and Oh, 2011). These species exhibit a wide range of morphological variation even among individuals within populations (Yu et al., 1981a; Choi and Oh, 2011). Choi and Oh (2011) distinguished the latter from the former by several morphological characters (clearly elongated leaf sheath, which is usually exposed above ground, and longer scape), and different chromosome numbers: A. thunbergii is a diploid with 2n = 16, whereas A. sacculiferum is a tetraploid with 2n = 32. However, Choi and Oh (2011) also noticed a diploid population that, based on morphological observations, belongs within the range of tetraploid A. sacculiferum.
The goals of this study are (1) to broaden the current knowledge on cytogeography and ecology (in addition to Choi and Oh, 2011) of A. thunbergii and A. sacculiferum, especially focused on the South Korean populations; (2) to examine whether diploid and tetraploid populations show segregation in relation to the elevational gradient or habitat types; and (3) to address taxonomic confusion between A. thunbergii and A. sacculiferum.
Materials and Methods
The collected samples were tentatively morphologically identified based of Choi and Oh (2011), and details of sampled populations are listed in Table 1. The coordinates and elevations were recorded with a Garmin GPS 60CSx instrument (Taipei, Taiwan). To determine chromosome numbers, shoot tips were pretreated in 0.002 M 8-hydroxyquinoline for 4–6 h in darkness at 4°C and then fixed in Carnoy’s fluid (3 parts absolute ethanol:1 part glacial acetic acid, v/v) for 1 h at room temperature (23°C). The shoot tips were macerated in 1M hydrochloric acid at 60°C for 10–15 s. After washing 3–5 times to eliminate residual hydrochloric acid and staining with 1% aceto-orcein for 8 h, the material was squashed for observation in 45% acetic acid. More than 10 micrographs were taken for each accession by an optical microscope (Olympus AX-70, Tykyo, Japan) (Fig. 1). Semipermanent microscope slides and photographs of representative cells have been retained in the Plant Systematics Laboratory of the Changwon National University, South Korea. We used Mann-Whitney U-test to test differences in elevation between di- and tetraploids.
Results
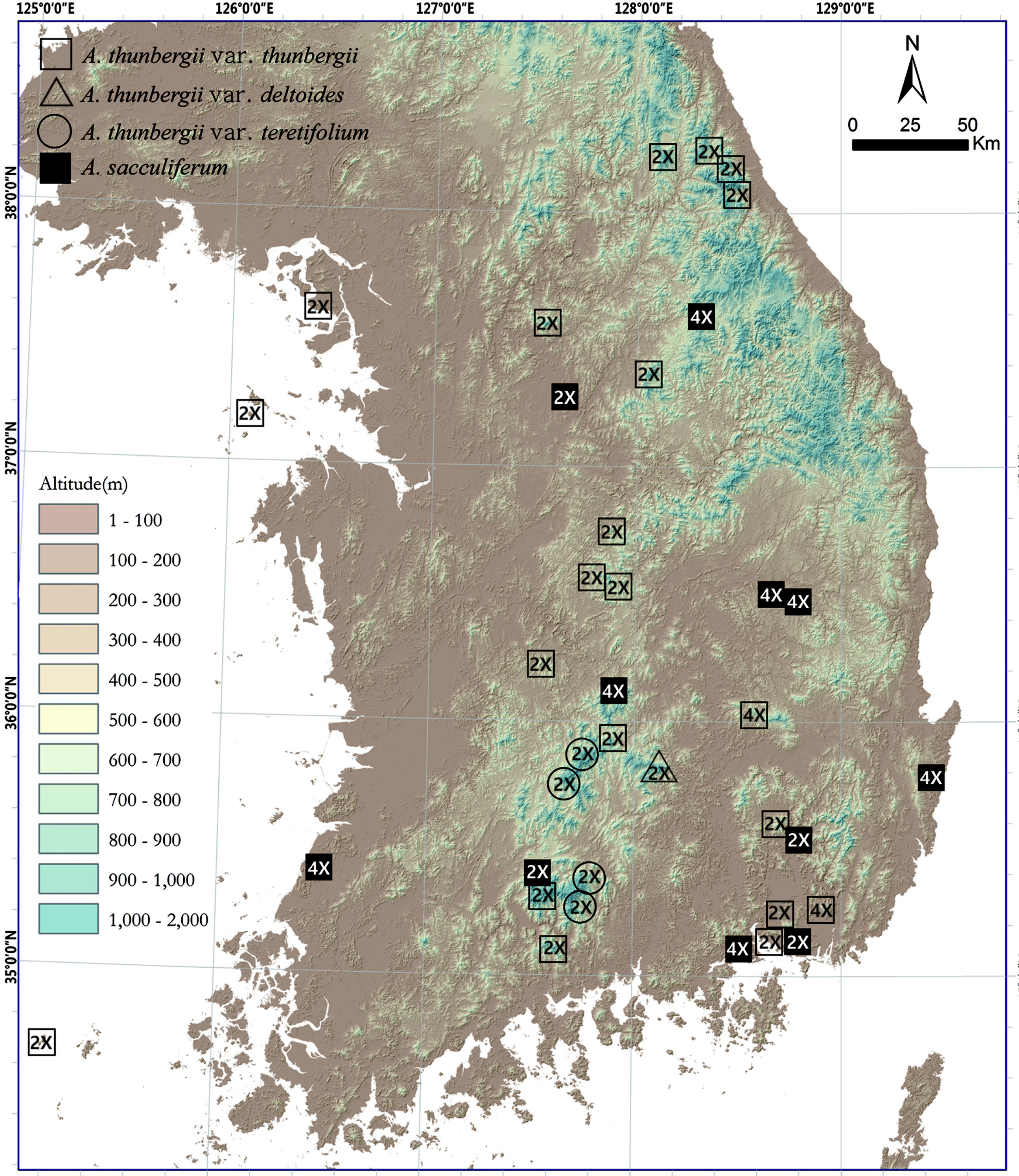
A total of 97 and 65 individuals from 26 and 13 populations of tentatively morphologically identified A. thunbergii (including var. thunbergii, var. deltoides, and var. teretifolium) and A. sacculiferum were counted for chromosome numbers, respectively (Fig. 1, Table 1). Only single-ploidy populations were found in both species (mean = 4.7 and 5 samples per population in A. thunbergii and A. sacculiferum, respectively) (Fig. 2, Table 1). The most frequent cytotype of A. thunbergii was diploid (2n = 16) found in 24 out of 26 populations (92.3%) while rare tetraploid cytotype (2n = 32) was found in remaining 2 populations. Most A. sacculiferum populations (69.2%) were tetraploid including two populations from eastern Russia and China and remaining populations were diploid (Table 1).
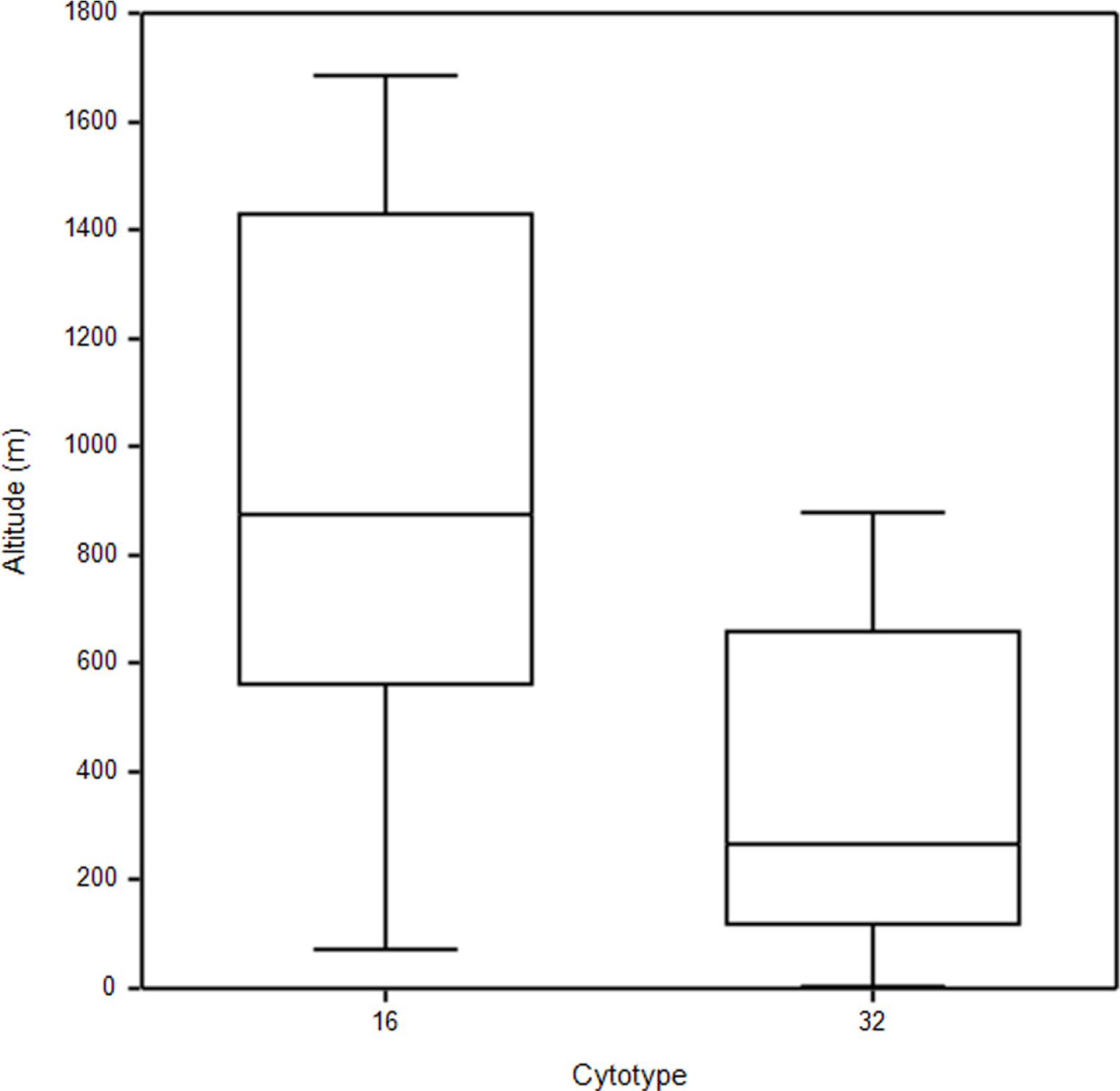
Most diploids of both species were found at higher elevations above ca 900 up to 1,700 m, whereas tetraploids of both species tend to occur below 800 m. However, some diploid populations were also found at relatively low elevations (Fig. 3). Median elevation of diploids (876 m) was significantly higher than that of tetraploids (268 m) (Mann-Whitney U-test, Z = −3.17, p = 0.002) (Fig. 3).
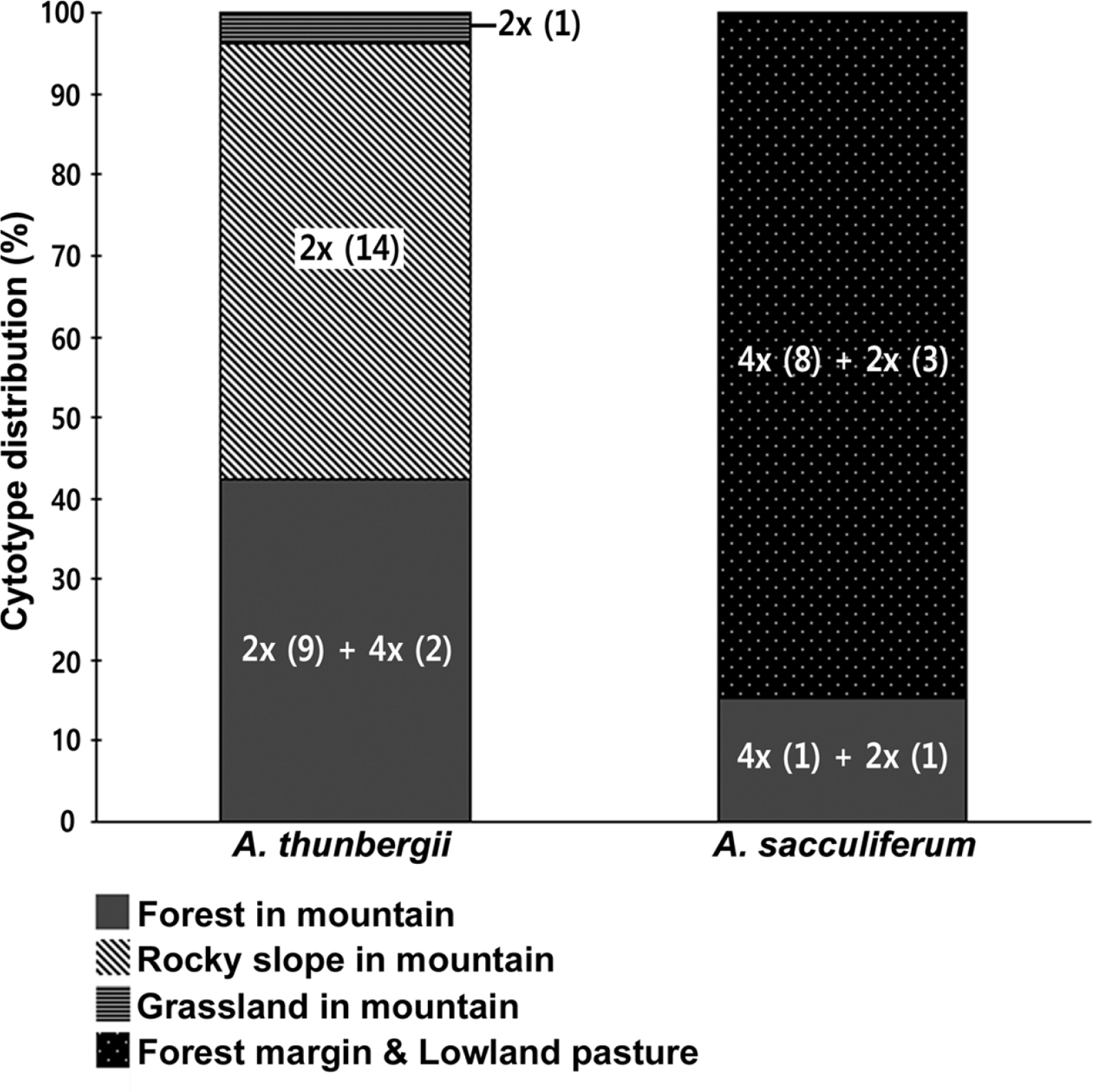
Habitat preferences of the cytotypes of two species were analyzed (Fig. 4). Allium sacculiferum occurs commonly in lowland meadows, forest margins and at riversides, while A. thunbergii usually occurs in rather different habitat types, such as sunny slopes of the rocky mountain and the forest in mountainous areas (Fig. 4). Out of 26 populations of A. thunbergii, nine diploids and two tetraploids occurred mountain forest (42.3%), 14 diploid populations occurred in the mountain rocky slopes (53.8%), and one diploid population was found in mountain grassland (3.8%) (Fig. 4). Whereas, out of 13 populations, one diploid and one tetraploid of A. sacculiferum grew in the forest in mountain (15.4%), and three diploids and eight tetraploids tend to occupy only in a forest margin and lowland pasture (84.6%) (Fig. 4).
Discussion
We observed two different ploidy levels (diploid and tetraploid) in both tentatively morphologically identified A. thunbergii and A. sacculiferum in various habitat conditions (Table 1). Compared to Choi and Oh (2011), tetraploids of A. thunbergii (Pop 16 and 18 in Table 1) were newly recorded in Korea. The main cytotype of A. thunbergii was diploid (92.3% of total). In contrast, the majority of A. sacculiferum was tetraploid (69.2% of total). Any population of the studied taxa did not harbor both cytotypes. In general, occurrence of triploids in sympatric 2x + 4x populations gives evidence of hybridization between diploid and tetraploid individuals while triploid detection in diploid populations indicates frequent production of unreduced gametes by diploids (Sanders, 1987; Noda and Kawano, 1998; Ramsey and Schemske, 1998). Absence of triploid record in our survey might be caused by either just extensive study with low number of analyzed plants or low probability of triploid production in diploids. However, from the representative karyotypes (Ko et al., 2009) (Fig. 1), we can present a possibility that the tetraploids of A. sacculiferum might be originated via autopolyploidization of diploid A. thunbergii or A. sacculiferum. Forthcoming studies by molecular cytogenetics as well as molecular sequencing data are required to more accurately identify the parental diploids genomes of the tetraploids.
Allium thunbergii was more often found at higher elevations than A. sacculiferum, and exhibited a tendency to occur more frequently in rocky slopes and in forests of mountainous areas. On the other hand, A. sacculiferum clearly preferred forest margins and lowland pastures (Choi and Oh, 2011) (Figs. 3, 4, Table 1). The cytotypes differed with respect to elevation; the diploids were found more frequently at higher elevations than the tetraploids (Fig. 3). It has been believed in past that cytotypes of higher ploidy level occur more frequently in higher elevations and/or are better colonizers of once glaciated areas (Lewis, 1980; Bierzichudek, 1985) due to their better physiological and ecological flexibility to adapt to harsh conditions. Concerning genus Allium, several studies examined the correlation of cytotypes with elevation in various species. For example, tetraploids of A. przewalskianum Regel occur mainly in higher elevations than diploids in the Qinghai-Tibetan Plateau and adjacent regions of China (Xie-Kui et al., 2008), and tetraploids of A. oleraceum L. mainly occur in higher elevations than the penta- and hexaploid individuals in Slovakia (Šafářová et al., 2011). However, Duchoslav et al. (2010) reported that there was no difference in elevation among ploidy levels of A. oleraceum in neighbouring region (the Czech Republic). This could be explained by different origins of tetraploids in each region and/or differences in migration routes of each cytotype even in geographically close regions. Elevational segregation of cytotypes with diploids occurring in higher elevations than tetraploids has previously been reported for Anthoxanthum spp. (Felber-Girard et al., 1996; Filipová and Krahulec, 2006), Chamerion angustifolium (L.) Holub (Husband and Schemske, 1998) and Senecio carniolicus Willd. (Schönswetter et al., 2007). Based on the results of this study, we can make a hypothesis on the phytogeography of Korean A. thunbergii complex (including A. sacculiferum) that firstly the diploid ancestor had settled along the higher elevational mountainous areas in natural habitats, and after polyploid formation tetraploids colonized various semi-natural habitats especially in lower elevational areas. However, additional wider range studies covering whole distribution range of A. thunbergii complex are required for the clear elucidation of its phytogeography in Korea.
Allium sacculiferum differs from A. thunbergii by minute morphology characters: length of the leaf and scape, cross section of leaf blades; A. sacculiferum has a clearly elongated leaf sheath, which is exposed above ground and a longer leaf and scape (Choi and Oh, 2011). In addition to the morphological identification, Ko et al. (2009) and Choi and Oh (2011) found that these closely related species are either diploid (A. thunbergii) or tetraploid (A. sacculiferum). Choi and Oh (2011) also noticed a diploid population that, based on morphological observations, belongs within the range of tetraploid A. sacculiferum. The morphological differentiation between tetraploids and diploids in several polyploid plant complexes is documented by several studies. Typically, polyploidy increases the sizes of cells (e.g., guard cells) and organs, and it can result in increase of more complex traits as well as overall habit (Müntzing, 1936; Otto and Whitton, 2000; Levin, 2002; Beaulieu et al., 2008). For instance, the study of Sugiyama (2005) on Lolium perenne L. and L. multiflorum Lam. and Gao et al. (2016) on Chrysanthemum lavandulifolium (Fisch. ex Trautv.) Makino showed that leaves and stem (length, width) of tetraploids were bigger than those of diploids. Larger size of several traits in tetraploid A. sacculiferum in contrast to diploid A. thunberii is consistent with the mentioned studies, and we hypothesize that the autotetraploid population of A. thunbergii showing lager size has been therefore recognized as A. sacculiferum. However, the diploids of A. sacculiferum (Pop 31, 32, 35 and 36 in Table 1) clearly had also elongated leaf sheath in four populations while some tetraploids (Pop 16 and 18 in Table 1) of A. thubergii had a non-elongated to intermediate leaf sheath (H. J. Choi, pers. obs.). This could be explained by the adaptation of the respective population to various environmental factors. Therefore, the morphological characteristics of A. thunbergii and A. sacculiferum might be influenced by local adaption and polyploidization.
Although we studied populations that were morphologically identified as A. thunbergii and A. sacculiferum only in restricted part of the potential distribution area of these species, result from this study and additional biosystematics data suggest that A. thunbergii and A. sacculiferum do not still fulfil any species concept: (1) they are incompletely differentiated by morphological characters (morphological species concept), and overlap in majority of morphological characteristics including reproductive organs in particular (Choi and Oh, 2011; H. J. Choi, pers. obs.); (2) habitat differentiation (ecological species concept) is more or less incomplete (Fig. 4); (3) incomplete reproductive isolation (biological species concept) was recognized by artificially making viable seeds crossing diploid A. thunbergii and diploid A. sacculifeum (Yu et al., 1981b); and (4) Choi et al. (2012) showed an incomplete monophyletic network of DNA sequences (phylogenetic species concept) between A. thunbergii and A. sacculiferum.
Plants with different chromosome numbers are not infrequently found within the bounds of one taxonomic species (Friesen, 1992; Rice et al., 2015). The problem of the taxonomic rank of the members of one polyploidy series of plants is so far disputable. Based on Friesen (1992), we took up the following position in understanding the species range in the genus Allium. In the cases when definite populations of plants are the members of the polyploidy series, and besides they constantly differ in morphological characters and have the habitat of their own, we can distinguish them as independent taxonomic species. Friesen (1992) also suggested that it is untimely to impart the rank of a taxonomic species to cytotypes with difference in the chromosome numbers but without revealing appreciable morphological differences. Consequently, A. thunbergii and A. sacculiferum should be regarded as a same species. Therefore, we propose to treat A. sacculifeum as an additional synonym of A. thunbergii. Additionally, Allium thunbergii G. Don var. deltoides (S. Yu. W. Lee & S. Lee) H. J. Choi & B. U. Oh is unified into the basic species of A. thunbergii var. thunbergii. Future studies involving various cases on the cytotype diversity and its taxonomic significance will provide a sound basis to understand more clearly the species boundary of Allium.
Taxonomic Treatment
Allium thunbergii G. Don., Mem. Wern. Soc. 6: 84, 1827.—TYPE: Japan. UPS 8065 (holotype: UPS!, Fig. 5).
A. odorum Thunb., Fl. Jap 132, 1784. Non L. Type not traced.
A. morrisonense Hayata., Icon. Pl. Formos. 6. Suppl: 84, 1917; A. bakeri Regel var. morrisonense (Hayata) T. S. Lui & S. S. Yung., Fl. Taiwan. 5: 45, 1978. Type not traced.
A. sacculiferum Maxim., Prim. Fl. Amur. 281, 1859.—TYPE: Russia. On south Amur, one Dai travel over the Chinganskoi Piket, 21 Aug 1856, K. I. Maximowicz. s. n. (lectotype: LE!, designated by N. Friesen, 8 Aug 1996, photo CBU!).
A. ophiopogon H. Lév., Repert. Spec. Nov. Regni Veg. 12: 184, 1913.—TYPE: Korea. Quelpaert, in Parva insula Mouseum, in herbidis, 9 Aug 1911, Taquet 5213 (lectotype: TI!).
A. stenodon Nakai & Kitag., Rep. 1st. Sci. Exp. Manch. 4: 18, 1934.—TYPE: Korea. In monte Wu-ling-shan, 2 Sep 1933, N. H. K. s. n. (holotype: TI?).
A. komarovianum Vved., Bull. Univ. As. Centr. 19: 119, 1934.—TYPE: Russia. By Village Faddeevka on the river Suifun, 11 Sep 1931, V. L. Komaov s. n. (holotype: LE!, photo: CBU!).
A. sacculiferum Maxim. var. viviparum Sakata, Bull. Nat. Sci. Mus. Tokyo 7: 16, 1938.—TYPE: Japan. Keiki, pede Reikistan, Suigen, 6 Oct 1935, T. Sakata s. n. (holotype: TI?).
A. deltoidefistulosum S. Yu, W. Lee & S. Lee, Korean J. Pl. Taxon. 11: 30, 1981.—TYPE: Korea. Jeonbuk: Namwon, wet open grassland around pine forest at 600 m c. 800 m toward Segeolsan of Gongan, Yoo 6012 (lectotype: KWNU!; isolectotypes: KWNU!, JNU?, SNU?).
A. yuchuanii Y. Z. Zhao & J. Y. Chao, Acta Sci. Nat. Univ. Intramongolicae 20: 241, 1989. Type not traced.
A. cyaneum Regel var. deltoides S. Yu, W. Lee & S. Lee, Korean J. Pl. Taxon. 11: 29, 1981.—TYPE: Korea. Gyeongnam: Gayasan, grassland and soil on the rocks c. 1,300– 1,400 m, along the trail to the peak, Yoo 5920 (holotype: Wonkwang University herbarium; isotypes: KWNU!, JNU, SNU); A. thunbergii G. Don var. deltoides (S. Yu., W. Lee & S. Lee) H. J. Choi & B. U. Oh, Korean J. Pl. Taxon. 33: 351, 2003.