The complete chloroplast genome of Glycyrrhiza uralensis Fisch. isolated in Korea (Fabaceae)
Article information
Abstract
The chloroplast genome of Glycyrrhiza uralensis Fisch was sequenced to investigate intraspecific variations on the chloroplast genome. Its length is 127,689 bp long (34.3% GC ratio) with atypical structure of chloroplast genome, which is congruent to those of Glycyrrhiza genus. It includes 110 genes (76 protein-coding genes, four rRNAs, and 30 tRNAs). Intronic region of ndhA presented the highest nucleotide diversity based on the six G. uralenesis chloroplast genomes. A total of 150 single nucleotide polymorphisms and 10 insertion and deletion (INDEL) regions were identified from the six G. uralensis chloroplast genomes. Phylogenetic trees show that the six chloroplast genomes of G. uralensis formed the two clades, requiring additional studies to understand it.
Glycyrrhiza uralensis Fisch. is distributed mainly in Central Asia, Mongolia, and China (Hayashi and Sudo, 2009). Together with two sister species, Glycyrrhiza glabra L. and Glycyrrhiza inflata Batalin, G. uralensis is one of important traditional Chinese medicine used as sweetener (Rizzato et al., 2017), and has long been cultivated in China and Korea (Hayashi and Sudo, 2009; Park et al., 2020a). Because G. uralensis is recognized as the best species among three Glycyrrhiza species with recent decease of production of G. uralenesis in inner Mongolia, the closely related species of G. uralensis have been considered as alternative species (Lee et al., 2010; Kim et al., 2019a). Due to this importance, multiple number of chloroplast genomes have been sequenced till now (Kang et al., 2018; Jia et al., 2019) and several molecular markers, such as simple sequence repeats, have been developed (Hayashi et al., 2005; Lee et al., 2019; Hantemirova et al., 2020). Moreover, various useful compounds, including antibacterial and antithrombotic compounds, have been identified from G. uralensis (He et al., 2006; Kwon et al., 2010; Lee et al., 2010; Tao et al., 2012; Adianti et al., 2014). In addition, natural hybrids of Glycyrrhiza were reported (Chen et al., 2020) and some cultivars through hybridization were developed (Lee et al., 2017). Here, we completed the chloroplast genome of G. uralensis cultivated in Korea to investigate intraspecific variations on chloroplast genomes and its intraspecific phylogeny.
Materials and Methods
Plant material
We collected the G. uralensis in Godo 9-gil, Iksan-si, Jeollabuk-do, in Korea (36o00′14.45″N, 127o03′49.47″E). A voucher and isolated DNA was deposited in the InfoBoss Cyber Herbarium (IN, the voucher number IB-01093).
DNA extraction and chloroplast genome determination
Total DNA was extracted from fresh root by using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using NovaSeq6000 at Macrogen Inc., Korea, resulting in 15.8 Gbp raw reads. De novo assembly was done by Velvet v1.2.10 (Zerbino and Birney, 2008) and GapCloser v1.12 (Zhao et al., 2011). Assembled sequences were modified and confirmed by BWA v0.7.17 (Li, 2013) and SAMtools v1.9 (Li et al., 2009). All analyses were conducted in the Genome Information System (GeIS; http://geis.infoboss.co.kr/) used in the previous studies (Bum et al., 2020; Kim et al., 2021).
Genome annotation was conducted based on another chloroplast genome of G. uralensis (MT120790) with Geneious Prime 2020.2.4 (Biomatters Ltd., Auckland, New Zealand). A circular map of G. uralensis chloroplast genome was drawn using OGDRAW v1.31 (Greiner et al., 2019).
Identification of intraspecific variations
Single nucleotide polymorphisms (SNPs) and insertions and deletions (INDELs) were identified from the multiple sequence alignment of the six G. uralensis chloroplast genomes conducted by MAFFT 7.450 (Katoh and Standley, 2013) with ‘Find variations/SNPs’ implemented in Geneious Prime 2020.2.4 (Biomatters Ltd.). In addition, multiple sequence alignments from eleven Glycyrrhiza species were used in the same way (Table 1). Moreover, pair-wise alignments of the selected chloroplast genomes (Table 1) were also utilized in the same way. This method has been used in the previous studies investigating intraspecific variations on organelle genomes (Choi et al., 2021; Park et al., 2021c, 2021d). INDEL region was defined as the continuous INDELs.
Nucleotide diversity analysis of chloroplast genomes
We calculated nucleotide diversity in the six chloroplast genomes of G. uralensis using the method proposed by Nei and Li (1979) with the Perl script. Nucleotide diversities were scanned along the genome with 500-bp window size and 200-bp step size for overlapped sliding windows. Our script for nucleotide diversity calculation has been utilized in the previous studies (Kim et al., 2019b; Lee et al., 2020a, 2020b, 2020c; Park et al., 2021e; Park and Xi, 2021).
Phylogenetic analysis
Maximum-likelihood (ML) and Bayesian inference (BI) phylogenetic trees were constructed based on the multiple sequence alignment of 57 Glycyrrhiza chloroplast genomes by MAFFT v7.450 (Katoh and Standley, 2013). The ML tree was reconstructed in MEGA X with 1,000 bootstrap repeats (Kumar et al., 2018). In the ML analysis, a heuristic search was used with nearest-neighbor interchange branch swapping, TVM + F + R4 model, and uniform rates among sites. All other options used the default settings. The posterior probability of each node was estimated by BI using MrBayes v3.2.6 (Huelsenbeck and Ronquist, 2001) plug-in implemented in Geneious Prime 2020.2.4 (Biomatters Ltd.). The HKY85 model with gamma rates was used as a molecular model. A Markov chain Monte Carlo algorithm was employed for 10,000,000 generations, sampling trees every 200 generations, with four chains running simultaneously. Trees from the first 2,500,000 generations were discarded as burn-in.
Results and Discussion
The chloroplast genome of G. uralensis (GenBank accession: MZ066516) isolated in Korea is 127,689 bp long, which is shorter than those of typical plant chloroplast genomes. This is caused by lack of one of inverted repeat regions in the typical plant chloroplast genomes (Fig. 1), which is a common phenomenon in genus Glycyrrhiza (Kang et al., 2018). However, Abrus chloroplast (GenBank accession: MT328396), neighbor genus of Glycyrrhiza (Hayashi et al., 1998), presents a typical structure of chloroplast genome, indicating that this atypical structure is specific to Glycyrrhiza. It contains 110 genes consisting of 76 protein-coding genes (PCGs), four ribosomal RNAs (rRNAs), and 30 transfer RNAs (tRNAs) (Fig. 1).

Circular map of chloroplast genome of Glycyrrhiza uralensis isolated in Korea. Gene shown outside are transcribed clockwise, and inside the circle are transcribed counter clockwise. Genes are color-coded to distinguish different functional groups. The dark grey and the light grey plot in the inner circle correspond to the GC content and AT content, respectively. Inside chloroplast genome, the three arcs of which colors are grey, light green, and light blue indicate large single copy (LSC), small single copy (SSC), and inverted repeat (IR) regions.
A total of 150 SNPs and 10 INDEL regions covering 75 bp were identified from the six G. uralensis chloroplast genomes. Eighteen out of 76 PCGs, including matK, atpB, ndhK, psaB, psbC, rpoB, rpoC1, rpoC2, atpF, atpA, rpl20, psbB, rps3, ycf2, ycf1, ndhL, ccsA, and ndhF, cover at least one SNPs. The number of PCGs containing at least one SNP is slightly larger than that of Chenopodium album L. (Park et al., 2021b); however, the ratio of non-synonymous to synonymous SNPs is 15:19, which is typical in anigosperms. The genes containing non-synonymous SNPs, matK, atpB, ndhK, rpoC1, rps3, ycf1, and ccsA, can be a candidate to develop Glycyrrhiza-specific molecular marker to evaluate their generic background.
To evaluate the numbers of SNPs and INDELs identified from G. uralensis chloroplast genomes, we investigated intraspecific variations of eleven Glycyrrhiza species (Table 1) using the available complete chloroplast genomes. The numbers of SNPs ranged from 11 to 1,163 and those of INDEL regions were from 8 to 281 (15 to 2,339 bp in length) (Fig. 2). Many of numbers of intraspecific variations of the other plant species showed similar or smaller than those of G. uralensis except Goodyera schlechtendaliana Rchb. F. (Fig. 2, Table 2). It indicates that the number of SNPs of G. uralensis is in the middle, but INDEL coverage is relatively small. However, genetic diversity of chloroplast genomes should be investigated further based on well-designed sampling strategy considering with the number of samples and their locations together.

Scattered plot of number of single nucleotide polymorphisms (SNPs) and insertion and deletion (INDEL) coverage (bp) identified from the twelve Glycyrrhiza species and nine species. X-axis is number of SNPs and Y-axis is INDEL coverage. Blue dot indicates the number of SNPs and INDEL coverage identified from G. uralensis, orange dots were from eleven Glycyrrhiza species, and grey dots were from those of nine plant species identified based on pair-wise alignment of complete chloroplast genomes.
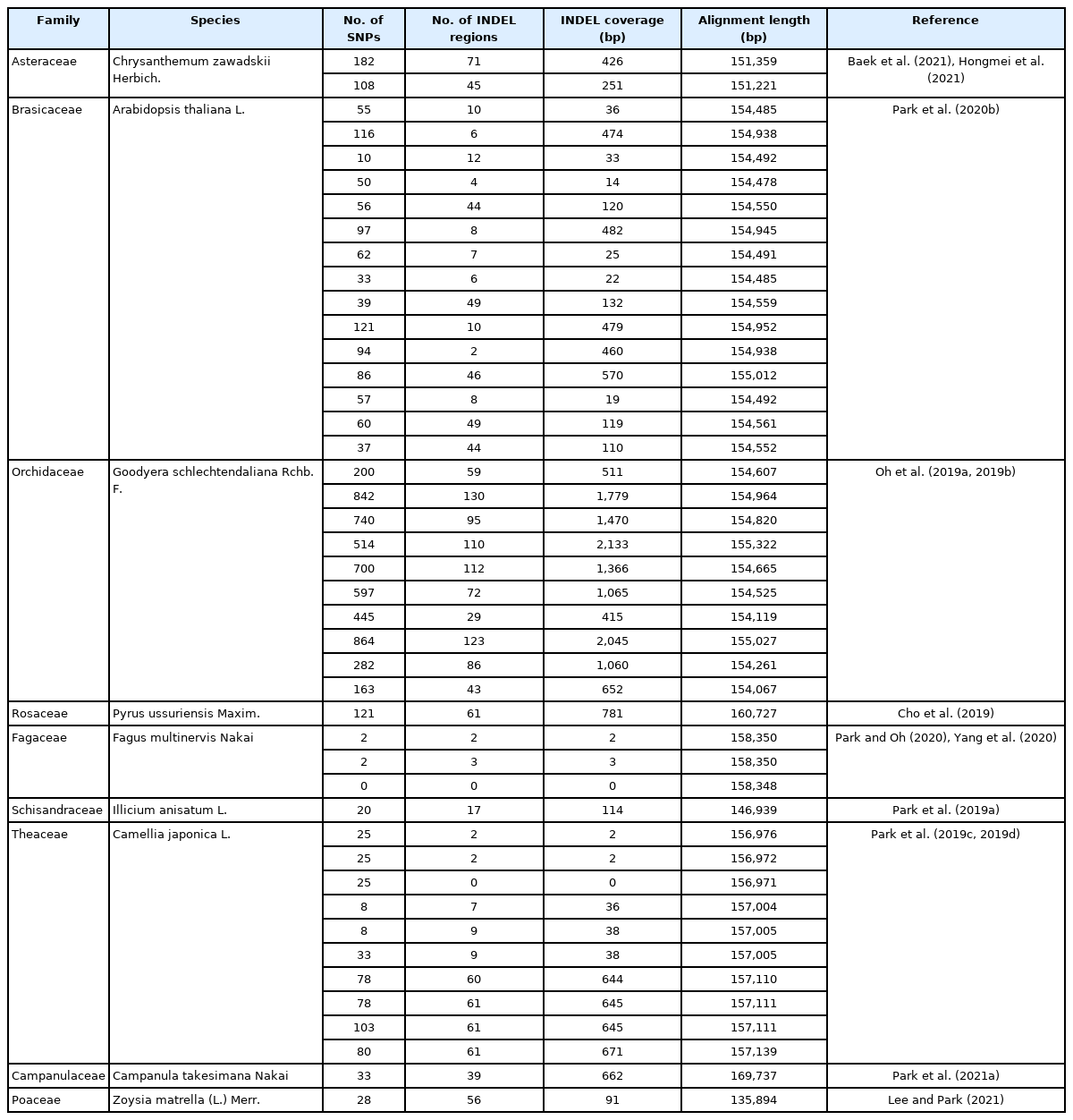
List of number of intraspecific variations identified from chloroplast genomes of the various plant species.
Nucleotide diversity of six G. uralensis chloroplast genomes was calculated to investigated hotspot of nucleotide diversity. Average nucleotide diversity was 0.0002216 (Fig. 3A) which is higher than those of A. thaliana (0.000017) isolated from several countries (Park et al., 2020b) and C. album (0.0000625) isolated in the Korean Peninsula (Park et al., 2021b). This high intraspecific nucleotide diveristy may be caused by different genetic backgrounds of G. uralensis, in which accessions are separated in two indepdendent clades in the phylogenetic tree (Fig. 3A). Interestingly 5′ region of atpB contained three non-synonymous SNPs (Fig. 3B), which is congruent to the studies presenting that atpB is one of positive selected genes (Xie et al., 2018; Lee and Park, 2021; Zhang et al., 2021). In addition, intron region of ndhA also displayed high nucleotide diversity, caused by one big insertion region from MT120791 (Fig. 3C). This phenomenon was also observed in various chloroplast genomes, such as A. thaliana (Park et al., 2020b), Viburnum amplificatum Kern (Park et al., 2020c), and Coffea arabica L. (Park et al., 2019b). Furthermore, intergenic regions of petN-trnC, trnC-rpoB, atpI-atpH, atpA-trnG, trnQ-accD, accD-psaI, psbE-petL, and trnR-trnL presented high nucleotide diversity (Fig. 3A), which can be used for developing molecular markers to distinguish G. uralensis populations or cultivars.
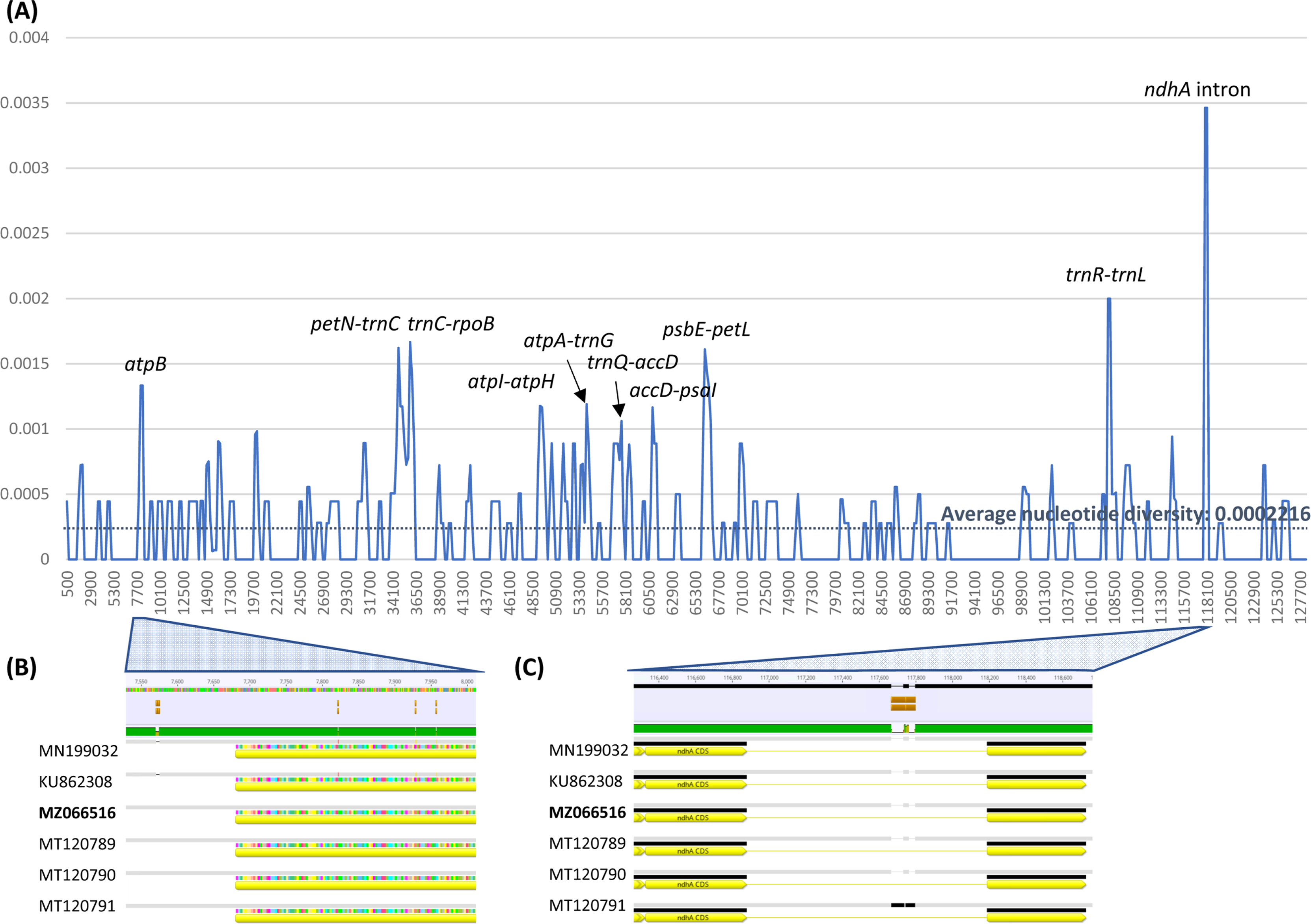
Nucleotide diversity calculated from the six G. uralensis chloroplast genomes. A. X-axis indicates coordination of chloroplast genome and Y-axis indicates nucleotide diversity of each window. Average nucleotide diversity was displayed as dark blue color and labels on graph indicate nearby gene names of the peak of which nucleotide diversity is above 0.001. B. Enlarged diagram shows multiple sequence alignment in 5′ region of atpB. C. Enlarged diagram shows multiple sequence alignment in 5′ region of ndhA intron.
Fifty-seven Glycyrrhiza chloroplast genomes with one outgroup species of Abrus pulchellus subsp. cantoniensis (Hance) Verdc., were used for constructing phylogenetic trees. Both phylogenetic trees display that six G. uralensis are clustered into the two clades including one G. glabra (GenBank accession: MG736059) with high supportive values of ML and BI trees except one low bootstrap value of ML (Fig. 4). Phylogenetic position of G. uralenesis is congruent to the previous phylogenetic studies (Hayashi et al., 1998, 2000; Liu et al., 2019; Duan et al., 2020). In addition, one G. glabra chloroplast (GenBank accession: MG736059) was found in the clade of G. uralensis (Fig. 4). One of possibility of this chloroplast can be misidentification because G. glabra was in the same clade with G. inflata in the previous studies (Hayashi et al., 1998, 2000; Liu et al., 2019; Duan et al., 2020). Moreover, two G. uralensis chloroplast genomes (GenBank accessions: KU862308 and MN199032) isolated in Korea and China (Jia et al., 2019), respectively, were clustered separately (Fig. 4), suggesting that geographical origin of G. uralensis might not be related to these two clades due to its cultivation in East-Asian countries (Yamamoto et al., 2003; Hayashi et al., 2005). Interestingly, non-synonymous SNPs identified only in the two G. uralensis chloroplast genomes (GenBank accessions: KU862308 and MN199032) were found in atpB (Fig. 3B) as well as ndhK, rpoC2, and ycf1. In addition, synonymous SNPs from those chloroplast genomes were also identified in psbA, psbC, rpoB, rpoC1, rpoC2, atpA, accD, psbB, and ndhF, which may contribute to forming the two separate clades. G. uralensis chloroplast genome assembled in this study provides insights of its intraspecific features, which will be helpful for understanding the structure of cultivated and native G. uralensis populations in detail.

Maximum-likelihood (ML) and Bayesian inference (BI) phylogenetic trees based on the complete chloroplast genomes of 57 Glycyrrhiza and one outgroup taxon, Abrus pulchellus subsp. cantoniensis. Phylogenetic tree was drawn based on ML tree. The numbers above branches indicate bootstrap values of ML tree and posterior probabilities of BI tree, respectively. Locations of each chloroplast genome were displayed after species name, originated from the published papers (Sabir et al., 2014; Kang et al., 2018; Bang et al., 2019; Jia et al., 2019; Duan et al., 2020) or source section of GenBank files of unpublished chloroplast genomes. Star mark in the NC_042146 indicates that location is not available and it is from the location of institute. The clade containing MZ066516 chloroplast was enlarged to make it easy to show the topology.
Acknowledgements
This study was carried out with the support of the Ministry of Small and Medium-sized Enterprises (SMEs) and Startups (MSS), Korea, under the “Regional Specialized Industry Development Plus Program (R&D, S2913418)” supervised by the Korea Institute for Advancement of Technology (KIAT).
Notes
Conflicts of Interest
The authors declare that there are no conflicts of interest.

