The complete chloroplast genome of Limonium tetragonum (Plumbaginaceae) isolated in Korea
Article information
Abstract
The chloroplast genome of Limonium tetragonum (Thunb.) Bullock, a halophytic species, was sequenced to understand genetic differences based on its geographical distribution. The cp genome of L. tetragonum was 154,689 bp long (GC ratio is 37.0%) and has four subregions: 84,572 bp of large single-copy (35.3%) and 12,813 bp of small single-copy (31.5%) regions were separated by 28,562 bp of inverted repeat (40.9%) regions. It contained 128 genes (83 protein-coding genes, eight rRNAs, and 37 tRNAs). Thirty-five single-nucleotide polymorphisms and 33 INDEL regions (88 bp in length) were identified. Maximum-likelihood and Bayesian inference phylogenetic trees showed that L. tetragonum formed a sister group with L. aureum, which is incongruent with certain previous studies, including a phylogenetic analysis.
Limonium, belonging to Plumbaginaceae family, is a cosmopolitan halophytic genus, and a few species inhabit alkaline soil away from the coastal area (Kubitzki, 1993; Morgan and Funnell, 2018). Limonium tetragonum (Thunb.) Bullock, distributed in coastal areas of Korea and Japan, is a biennial halophytic species with radical leaves (8–15 × 1.5–3 cm) and yellow corolla (Owhi, 1965; Park, 2007; Lee et al., 2011; Park et al., 2020a). The crude extracts and solvent-partitioned fractions of whole plants of L. tetragonum display antioxidant (Lee et al., 2011) and anti-cancer activities (Kong et al., 2008); ethyl acetate soluble fraction of aerial parts exhibits anti-liver fibrosis (Kim et al., 2016); that of whole plants shows anti-alcohol toxicity (Kim et al., 2015); and its methanol extracts of whole plants present hepatoprotective activities (Yang et al., 2014). We sequenced L. tetragonum from the western coastal area of Korea for investigating intraspecific variations on chloroplast genomes with chloroplast of L. tetragonum isolated in eastern seashore of Korea.
Materials and Methods
Plant material
We collected L. tetragonum in Aphaedo island in Shinan-gun, Jeollanam-do, Korea (34.8612N, 126.32629E). A voucher specimen and genomic DNA were deposited in the InfoBoss Cyber Herbarium (IN, the voucher number IB-00899).
DNA extraction and chloroplast genome determination
The total genomic DNA was extracted from fresh leaf by using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using HiSeqX at Macrogen Inc., Korea, and de novo assembly was done by Velvet v1.2.10 (Zerbino and Birney, 2008) and GapCloser v1.12 (Zhao et al., 2011). Assembled sequences were confirmed by BWA v0.7.17 (Li, 2013) and SAMtools v1.9 (Li et al., 2009). All bioinformatic analyses were conducted in the Genome Information System (http://geis.infoboss.co.kr/) utilized in the previous studies (Kim et al., 2018, 2019a, 2019b, 2019c, 2021; Bum et al., 2020; Park et al., 2021c).
Genome annotation was conducted based on another L. tetragonum chloroplast (MW085088.1) with Geneious R11 v11.0.5 (Biomatters Ltd, Auckland, New Zealand). A circular map of L. tetragonum chloroplast genome was drawn using OGDRAW v1.31 (Greiner et al., 2019).
Identification of intraspecific variations
Single nucleotide polymorphisms (SNPs) and insertions and deletions (INDELs) were identified using the ‘Find variations/SNPs’ function implemented in the Geneious R11 v11.0.5 (Biomatters Ltd, Auckland, New Zealand) based on the pairwise alignment of the two chloroplast genomes of L. tetragonum conducted by MAFFT 7.450 (Katoh and Standley, 2013). This method has been used in the previous studies (Kim et al., 2019a; Min et al., 2019a; Choi et al., 2021; Park et al., 2021b, 2021d). INDEL region was defined as the continuous INDELs.
Phylogenetic analysis
Maximum-likelihood (ML) and Bayesian inference (BI) phylogenetic trees were constructed based on the multiple sequence alignment of all available nine Plumbaginaceae chloroplast genomes and the seven chloroplast genomes of non-core Caryophyllales clade (Crawly and Hilu, 2012; Yao et al., 2019) by MAFFT v7.450 (Katoh and Standley, 2013). The chloroplast genome of Nepenthes graciliflora Elmer (1912) (Yao et al., 2019) was marked as outgroup species. The ML tree was reconstructed in IQ-TREE v1.6.12 (Nguyen et al., 2015) with 1,000 bootstrap repeats. In the ML analysis, a heuristic search was used with nearest-neighbor interchange branch swapping, GTR+F + R4 model determined as the best fit model by the ModelFinder implemented in IQ-TREE, and uniform rates among sites. All other options used the default settings. The posterior probability of each node was estimated by the BI using MrBayes v3.2.6 (Huelsenbeck and Ronquist 2001). The HKY85 model with gamma rates was used as a molecular model. A Markov chain Monte Carlo algorithm was employed for 1,100,000 generations, sampling trees every 200 generations, with four chains running simultaneously. Trees from the first 100,000 generations were discarded as burn-in.
Data availablity
Chloroplast genome sequence can be accessed via accession number of MN044572 in GenBank of NCBI at https://www.ncbi.nlm.nih.gov. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA737051, SAMN19678692, and SRR14793527, respectively.
Results and Discussion
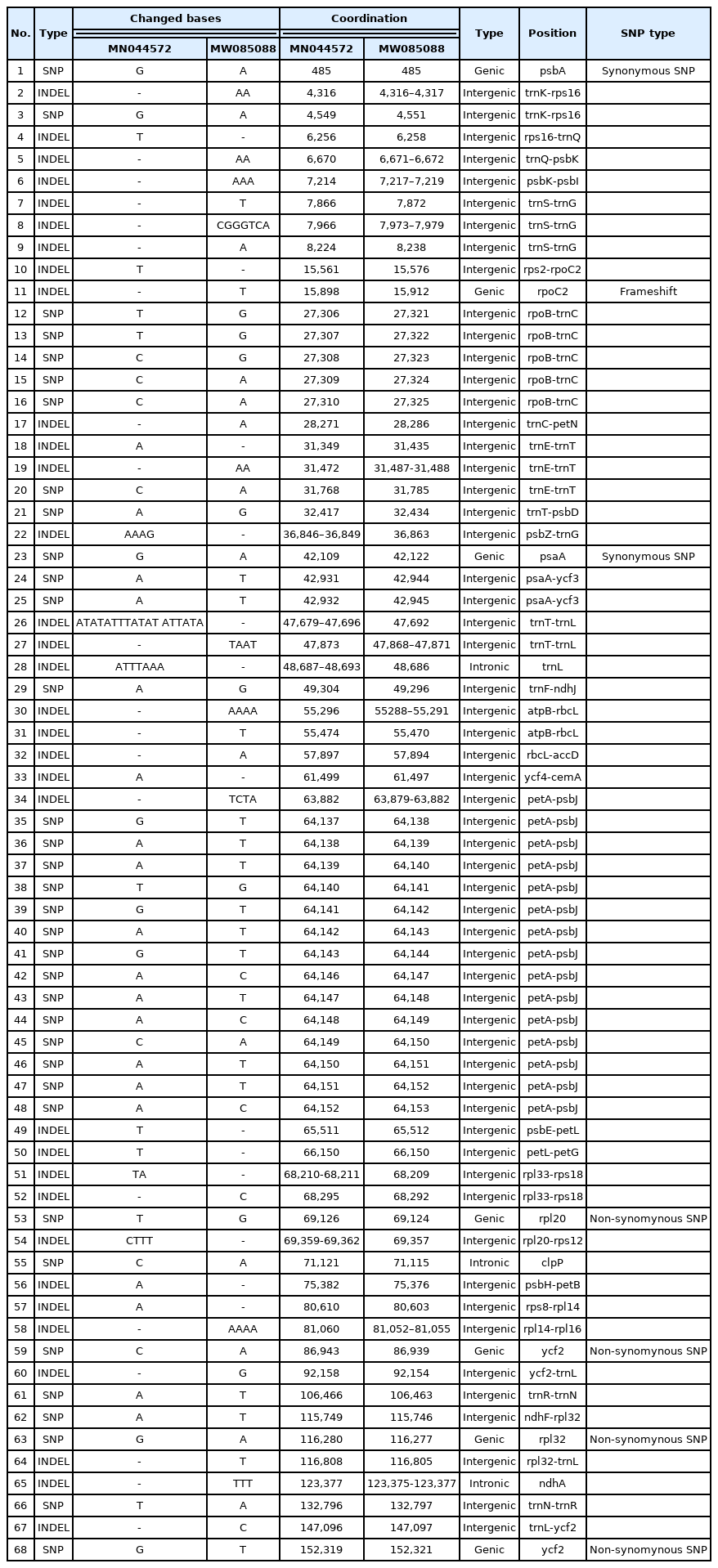
The chloroplast genome of L. tetragonum (GenBank accession of MN044572) isolated in Korea is 154,689 bp long (GC ratio of 37.0%) and had four subregions: 84,572 bp of large single copy (35.3%) and 12,813 bp of small single copy (31.5%) regions are separated by 28,562 bp of inverted repeat (IR; 40.9%) (Table 1), which is similar to those of the previously reported L. tetragonum chloroplast genome (MW085088) (Table 1). It contained 128 genes (83 protein-coding genes [PCGs], eight rRNAs, and 37 tRNAs); 14 genes (five PCGs, four rRNAs, and five tRNAs) were duplicated in the IR regions, which is also same to those of the previously reported L. tetragonum chloroplast genome (MW085088) (Fig. 1, Table 1).
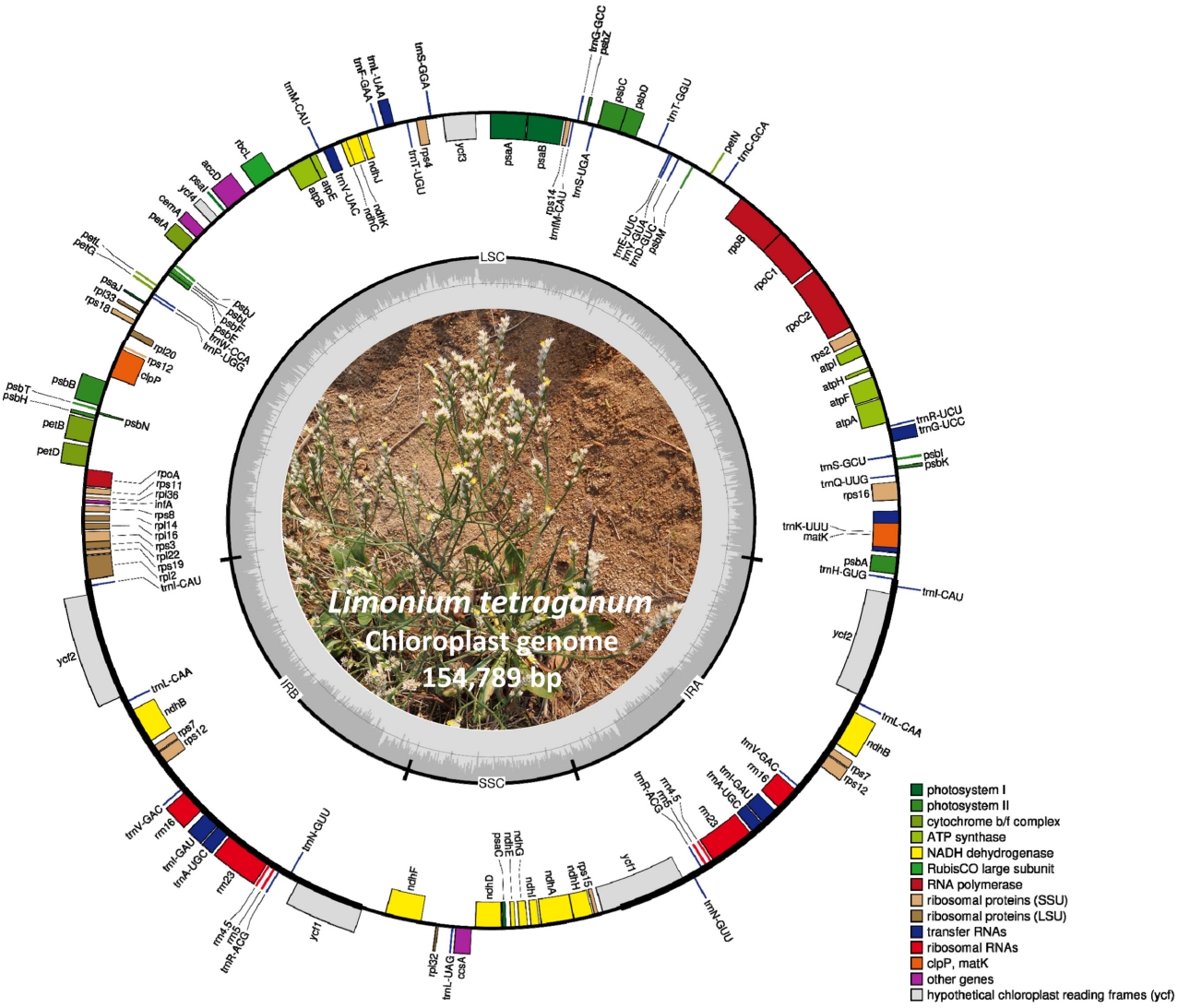
Circular map of chloroplast genome of Limonium tetragonum isolated in Korea. Genes shown outside are transcribed clockwise, and those inside the circle are transcribed counter clockwise. Genes are color-coded to distinguish different functional groups. The dark grey and the light grey plot in the inner circle correspond to the GC content and AT content, respectively.
Based on the pair-wise sequence alignment with the previously reported L. tetragonum chloroplast genome (MW085088) (Darshetkar et al., 2021), 35 SNPs and 33 INDELs regions (88 bp in total) were identified. The longest INDEL region was 18-bp located between trnT and trnL. The inserted sequence was 18-bp repetitive sequences. In addition, two chloroplast genomes of Limonium bicolor (Bunge) Kuntze (Darshetkar et al., 2021) display extremely divergent manner, no SNP and 116 INDEL regions (15,840 bp in length), which is similar to the case of Phedimus takesimensis chloroplast genomes (129 SNPs and 112 INDEL regions (8,506 bp in length) (Park et al., in preparation).
Numbers of these intraspecific variations of L. tetragonum were smaller than those of Pseudostellaria palibiniana (Takeda) Ohwi (84 SNPs and 125-bp INDELs), Pyrus ussuriensis Maxim. (1,221 SNPs and 781-bp INDELs), Goodyera schlechtendaliana Rchb. f. (200 SNPs and 511-bp INDELs), Gastrodia elata Blume (324 SNPs and 630-bp INDELs) isolated in Korea; while they were larger than those of Artemisia fukudo Makino (seven SNPs and 12-bp INDELs) (Min et al., 2019b), Fagus multinervis Nakai (two SNPs and 2-bp INDELs) (Park and Oh, 2020), Aconitum coreanum (H. Lév.) Rapaics (five to 19 SNPs and 52-bp to 950bp INDELs) (Kim et al., 2019d), Viburnum erosum Thunb. (16 SNPs and 50-bp INDELs) (Choi et al., 2020), and Veronica nakaiana Ohwi (seven SNPs and 4-bp INDELs) (Lee et al., 2021). In addition, seven PCGs contained at least one SNP or INDELs, among which two synonymous SNPs were found in psbA and psaA (Table 2) and four non-synonymous SNPs were identified in rpl20, rpl32, and ycf2 located in the IR region (Table 2). The ratio of synonymous to non-synonymous SNPs was 0.5, which is different from the normal ratio, like Chenopodium album L. (Park et al., 2021a). One INDEL region was identified in the genic region of rpoC2, resulting the extension of rpoC2 length in comparison to the previously sequenced chloroplast genome (MW085088) (Table 2).
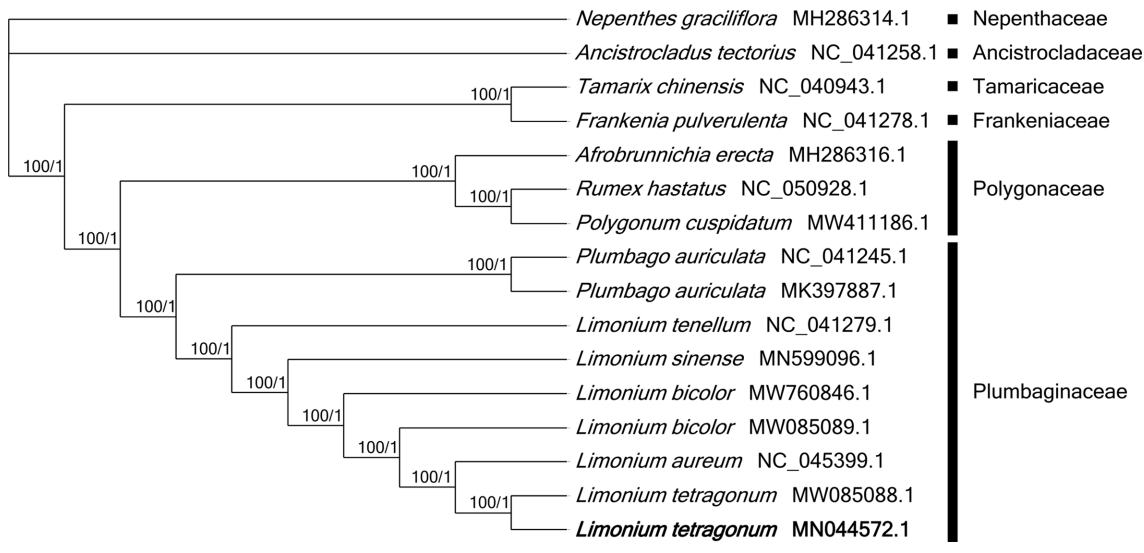
Sixteen chloroplast genomes were used for constructing ML and BI phylogenetic trees. Both phylogenetic trees displayed that two L. tetragonum, collected from the East and West coastal areas of Korean peninsula, respectively, were clustered with high bootstrap value (Fig. 2). The sister species of L. tetragonum is controversial: L. tetragonum formed a sister group with Limonium aureum (L.) Mill. in both trees (Fig. 2), whereas it was clustered with L. bicolor in the previous study using chloroplast genomes (Darshetkar et al., 2021). The close relative of L. tetragonum was not clearly confirmed because the phylogenetic study using chloroplast and nuclear loci could not determine the relationship with Limonium sinense (Girard) Kuntze, Limonium tenellum (Turczaninow) Kuntze, and Limonium flexuosum (Linnaeus) Kuntze (Koutroumpa et al., 2018) and the study with the complete chloroplast genome of L. tetragonum displayed that L. bicolor was sister species (Darshetkar et al., 2021), however, number of taxa used in this study was limited due to lack of complete chloroplast genomes and L. bicolor was not included in the previous study. This incongruency of the phylogenetic relationship might be caused by different sequences in both studies. Moreover, the supportive value of the clade of L. tetragonum and L. bicolor was 73%, which was lower than those (100%) in this study (Fig. 2), suggesting that L. aureum might be a sister species of L. tetragonum. Most of Limonium phylogenetic studies were conducted with the lack of East Asian Limonium species (Lledó et al., 2005; Malekmohammadi et al., 2017; Koutroumpa et al., 2018), requiring the additional phylogenetic studies including East Asian Limonium species.
Acknowledgements
This study was carried out with the support of the Honam National Institute of Biological Resources (HNIBR202101107).
Notes
Conflict of Interest
The authors declare that there are no conflicts of interest.