Phylogenetic position of Carex splendentissima, a Korean endemic sedge (Cyperaceae)
Article information
Abstract
Carex splendentissima U. Kang & J. M. Chung, endemic to the Korean peninsula, is characterized by staminate terminal spikes and glabrous elliptic perigynia. Based on its broad leaves, androgynous spikes, and tri-stigmatic features, the species has been placed in Carex sect. Siderostictae Franch. ex Ohwi, an East Asian section and a basal group in the genus. To clarify the monophyly and phylogenetic position of the species, a molecular study using the internal transcribed spacer region of nuclear ribosomal DNA and chloroplast DNA (trnL-F) data was conducted. The DNA sequence data of ten taxa in sect. Siderostictae and closely related taxa (two taxa in sect. Surculosae) with outgroups were analyzed based on maximum parsimony and maximum likelihood (ML) criteria. In the analyses, C. splendentissima was monophyletic and placed within the Siderostictae clade (sect. Siderostictae + two species of sect. Surculosae), forming a clade with C. ciliatomarginata and C. pachygyna (endemic to Japan). The clade (C. splendentissima + C. ciliatomarginata + C. pachygyna) shows evidence of diploidy. Furthermore, C. splendentissima is a sister to C. ciliatomarginata in the ML tree, and the two taxa have staminate terminal spikes. This study also updates the distribution of C. splendentissima and provides keys to the four Korean taxa in sect. Siderostictae. To conserve the endemic species C. splendentissima, further research on its genetic and ecological features should be conducted at the population level.
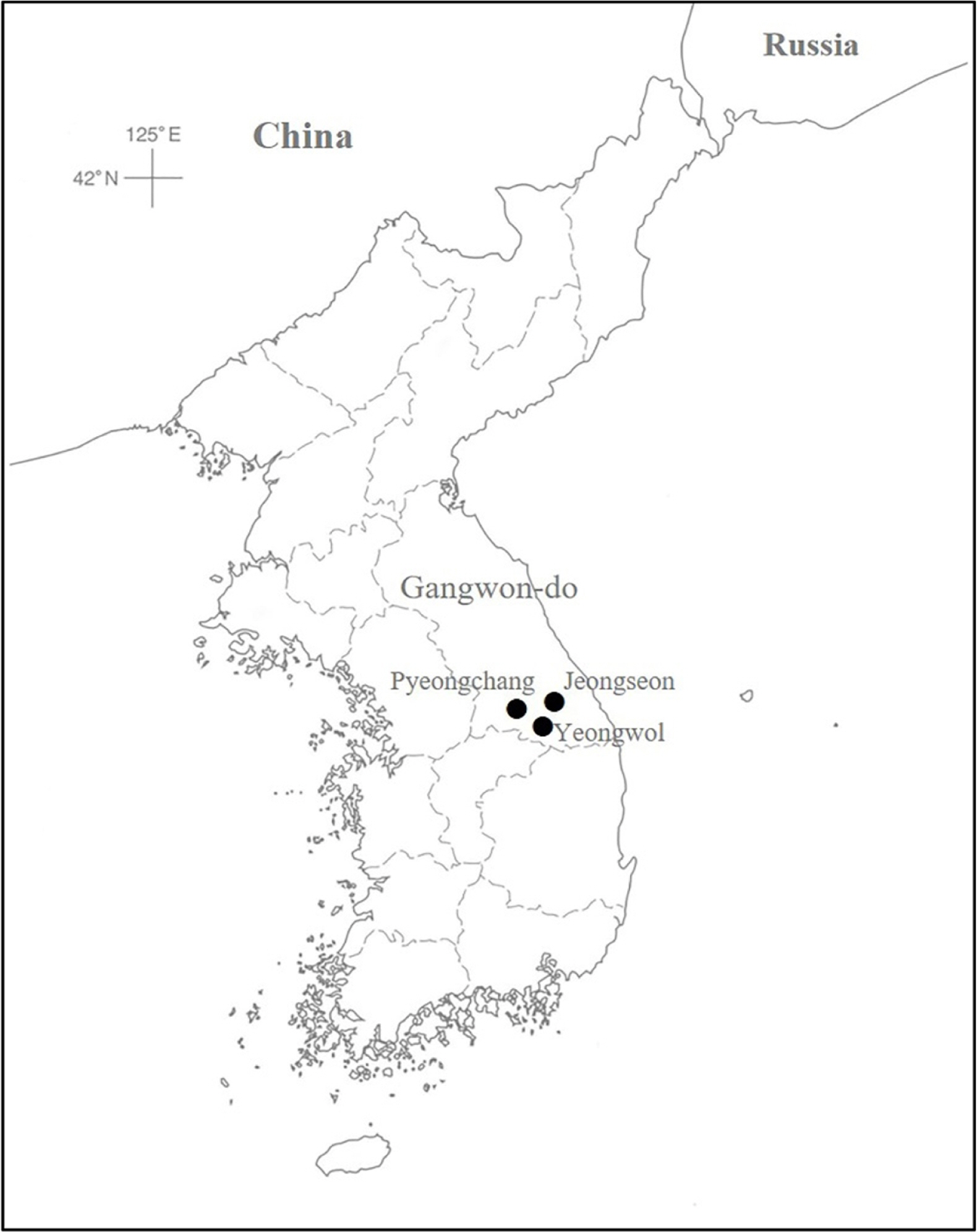
Carex splendentissima U. Kang & J. M. Chung (Cyperaceae) is an endemic to the Korean peninsula and distinguished by long rhizomes, glabrous leaves and perigynia, staminate terminal spikes, and tri-stigmas (Fig. 1) (Kang et al., 2012). It occurs only in Gangwon-do Province, mountainous areas in the middle east portion of the peninsula, on limestone rocks near streams (Figs. 1, 2) (Park et al., 2016). Based on broad leaves and bisexual lateral inflorescences (androgynous, staminate flowers at the tip and pistillate flowers at the base), Kang et al. (2012) placed the species in Carex L. sect. Siderostictae Franch. ex Ohwi (Carex subgenus Carex). The section occurs in East Asia in such countries as Japan, Korea, China, Vietnam, and Russia. A few species grow broadly in East Asia, but many taxa in the section are geographically structured, being endemic to certain country and/or areas (Table 1) (Egorova, 1999; Tang et al., 2010; Hoshino et al., 2011; Park et al., 2016). Among four taxa in Korea, two species are endemic: C. splendentissima and C. okamotoi Ohwi (Park et al., 2016).

Photographs of Carex splendentissima in a natural habitat (Jeongseon-gun, Gangwon-do). A, B. Habitats near streams. C. Populations associated with Saussurea chabyoungsanica and Aruncus dioicus var. kamtschaticus. D. Habits showing glabrous broad leaves. E. Inflorescence with terminal staminate and lateral androgynous spikes (staminate at the top and pistillate at the base).
Section Siderostictae is a very important group in a phylogenetic perspective being the earliest diverging group within the Cyperaceae tribe Cariceae (Waterway and Starr, 2007; Waterway et al., 2009). Because the East Asian section is basal to the rest of Carex species, to understand high diversity in Carex, the most specious genus in temperate zones with more than 2,000 taxa worldwide, it is critical to clarify evolutionary mechanisms and/or speciation in Asian groups (Global Carex Group, 2015). High diversity in Carex has been postulated by chromosome, genetic rearrangements via fission and/or fusion events in holocentric chromosomes, possessing non-localized centromeres (Hipp et al., 2009, 2013). Some previous investigations such as Chung et al. (2011) and Escudero et al. (2012) demonstrated that great variations in chromosome numbers are associated with rapid species diversity due to agmatoploidy (chromosome number increases by fission without gene duplications) (Luceño and Guerra, 1996) and/or aneuploidy in various linages. However, polyploidy events are very rare, and are only detected in sect. Siderostictae (Waterway et al., 2009). Polyploidy contributes to early lineage diversity whereas agmatoploidy and/or aneuploidy play an important role in recent diverging lineages in Carex (Waterway et al., 2009; Yano et al., 2014).
Within the section, terminal spike sexuality, perigynium shapes and sizes, distribution areas, and ploidy levels are various (Table 1). The basal phylogenetic position of the section indicates primitive character states of morphological, cytological, and geographic characters (Global Carex Group, 2015). In the analyses of the internal transcribed spacer region and 5.8S gene of nuclear ribosomal DNA (hereafter, ITS) and trnL intron and trnL-F intergenic spacer of chloroplast DNA sequences (hereafter, trnL-F), traditional sect. Siderostictae formed the Siderostictae clade with two sections Carex subgenus Vigneastra, sections Hemiscaposae C. B. Clarke and Surculosae Raymond, which are previously classified in (Yano et al., 2014). Chromosomes in these sections are larger in size and lower in number than in recently derived lineages (Waterway et al., 2009; Chung et al., 2013). Tetraploidy in the clade has been observed in the species occurring in relatively broader geographic areas, for examples, C. glossostigma Hand.-Mazz., C. grandiligulata Kük., and C. siderosticta Hance (Table 1) (Yano et al., 2014). Both Korean endemics, C. splendentissima and C. okamotoi, are diploids (2n = 12) (Chung et al., 2013, 2017).
To clarify monophyly and the phylogenetic position of C. splendentissima, molecular phylogenetic analyses were conducted using ITS and trnL-F sequence data. Furthermore, distribution of the species was updated based on voucher specimens, and keys to the four Korean members in sect. Siderostictae were presented.
Materials and Methods
Plant leaves were sampled from herbarium specimens or living plants at the Korea National Arboretum herbarium (KH), National Institute of Biological Resources herbarium (KB), and through field work at Jeongseon-gun, Gangwon-do Province. Voucher specimen information and accession numbers of DNA data are provided in Appendixes 1 and 2. Sequence data of some ingroups and outgroups were obtained from GenBank (Appendix 2). Total DNA were extracted from leaves using DNeasy Plant Mini Kit (Qiagen, Valencia, California, USA). ITS and trnL-F regions were amplified by polymerase chain reaction (PCR) using (Solg Taq DNA polymerase, SolGent Co., Daejeon, Korea). ITS-1 (Urbatsch et al., 2000) and ITS4 (White et al., 1990) primers were utilized for the ITS region, and trnL-F region primers followed Tablet et al. (1991). PCR reaction conditions were modified from Yano et al. (2014). PCR products were checked in 1% agarose gels, and then purified PCR products were sequenced in SolGent Co.
Raw sequence data were edited in Sequencher (version 4.8, Gene Codes Corporation, Ann Arbor, MI, USA) and aligned using MUSCLE (Edgar, 2004). Gaps were not used as characters, and tree searches were performed using parsimony and maximum likelihood (ML) methods. Most maximum parsimonious trees were obtained by PAUP* (Swofford, 2002) using the heuristic search option with 100 random addition sequence, tree-bisection-reconnection (TBR) branch swapping, and accelerated transformation (ACCTRAN) character-state optimization. RAxML v. 8 (Stamatakis, 2014) was used for the ML analyses using the GTRGAMAMA model, which was strongly supported (Chung et al., 2012). Tree searching for ITS and trnL-F data was conducted separately, and then a combined data set was analyzed. Prior to the analyses of combined data, the incongruence length difference (ILD) test evaluated ITS and trnL-F data for congruence with the heuristic searchers using random-sequence addition replicates in PAUP* (Farris et al., 1994, 1995). Branch support values were estimated by 1,000 heuristic bootstrap replicates with simple sequence addition and TBR branch swapping (Felsenstein, 1985).
Results and Discussion
Statistics of ITS, trnL-F, and combined data were summarized in Table 2. The combined 1,688 bp DNA sequence data of ITS (636 bp) and trnL-F (1,052 bp) were aligned and analyzed under maximum parsimony (MP) and ML assumptions. Number of parsimony-informative sites were higher in ITS (18%) than in chloroplast trnL-F data (6%). The ITS data resulted in 203 parsimony trees, and the trnL-F data produced 264 parsimony trees. Consistency Index (CI) and Retention Index (RI) were higher in the trnL-F than in the ITS data. ILD test p-value (0.04) allowed to conduct combined data analyses of the two data sets. The combined data of CI and RI were 0.7220 and 0.7540, respectively.
Both MP and ML analyses supported monophyly of C. splendentissima and placed it in the Siderostictae clade. C. splendentissima was included in the Siderostictae clade with sections Siderostictae and Surculosae and formed a clade with C. ciliatomarginata and C. pachygyna in sect. Siderostictae in all analyses. One of parsimony trees and an ML tree obtained from combined data set are presented in Fig. 3. Because all the parsimony trees and a strict consensus tree have similar topologies on the relationships and positions of C. splendentissima, one of parsimony tree is presented. The monophyly of C. splendentissima was strongly supported with high bootstrap values (≥ 99%) in MP and ML analyses. However, sister species were different between the two trees. C. ciliatomarginata was sister to C. splendentissima in ML analyses, while C. pachygyna in the maximum parsimony tree. The results supported the treatment of the species in sect. Siderostictae by Kang et al. (2012). Because of broad leaves and staminate terminal spikes, the species was often compared with C. ciliatomarginata in Korea (Park et al., 2016). C. okamotoi, the other Korean endemic, was nested with C. siderostacta with a high bootstrap value (≥ 80%). C. siderostacta is the most widely distributed species in the section. In Korea, the species grows commonly throughout the country, but C. okamotoi occurs mainly in Southern areas of the peninsula (Park et al., 2016). Diploidy has been reported in C. okamotoi (2n = 12), whereas diploidy and tetraploidy have been reported in C. siderostacta (2n = 12, 24) (Table 1). Polyploidy events in C. siderosticta may have caused complex genetic relationships. Further investigations with broad taxon and gene sampling are required to resolve the relationship between C. siderosticata and C. okamotoi. The analyses suggest that two Korean endemics, C. splendentissima and C. okamotoi, have evolved independently in the peninsula.

Phylogenetic trees of combined internal transcribed spacer and trnL-F data. A. One of parsimony trees (tree length = 552, Consistency Index = 0.7220, Retention Index = 0.7540). B. Maximum likelihood tree. Numbers on branches indicate bootstrap values more than 69%. In both analyses, Carex splendentissima forms a monophyletic group (dotted box) and the clade with C. pachygyna and C. ciliatomarginata within the Siderostictae clade.
The clade of C. splendentissima, C. ciliatomarginata, and C. pachygyna is diploidy (Table 1, Fig. 3). Two members of the clade, C. splendentissima (Korean endemic) and C. pachygyna (Japanese endemic), exhibit a narrow distribution pattern, supporting the association of distribution patterns with ploidy levels in Carex (Yano et al., 2014). Genome sizes (1C) of C. splendentissima and C. ciliatomarginata are 0.643 pg DNA and 0.742 pg DNA, respectively (unpublished data) whereas C. siderosticta, tetraploidy (2n = 24), has 1C = 1.20 pg DNA (Nishikawa et al., 1984). Polyploidization is significant in the clade of C. siderosticta and C. okamotoi, whereas C. splendentissima is in the diploid clade. Contrasting to the basal lineage (Siderostictae clade), C. scoparia var. scoparia in C. subgenus Vignea sect. Ovales has low genome size, 1C = 0.342–0.361 pg DNA (Chung et al., 2011). Genome sizes and chromosome numbers are not associated in the subgenus (Chung et al., 2012). Dynamic mechanisms of holocentric chromosomes, such as polyploidization in early lineages and agmatoploidy in recently diverging lineages, have contributed complex, diverse evolutionary history in the genus. The evolution of unisexual and bisexual spikes is challenging to understand in Carex. In Carex, unisexual inflorescences are diverse in sexuality, branching patterns, and positions; and the pistillate spikes with tri-stigmatic gynoecia and closed perigynia are present in early diverging lineages (Starr and Ford, 2009). In the Siderostictae clade, two types of terminal spike arrangements (staminate and androgynous) and two levels of ploidy levels (diploidy and tetraploidy) are present. Both characters are very important to understand evolution of Carex. Evaluation of the character evolution should be conducted with robust, well-resolved phylogenetic trees of the Siderostictae clade.
C. splendentissima is a specialist in ecological habitat preference, growing mainly on limestones. The distribution of the species was updated based on the voucher specimens examined in KB and KH (Fig. 2). In the Jeongseon habitat, the species was associated with other limestone lovers and/or ecological generalists such as Saussurea chabyoungsanica H. T. Im, Spiraea trichocarpa Nakai, Aruncus dioicus var. kamtschaticus (Maxim.) H. Hara, Polystichum sp., Lespedeza maximowiczii C. K. Schneid., and Lindera obtusiloba Blume. Limestone areas cover about 20% of South Korea mainly in middle East of the country (Korea National Arboretum, 2012). Limestone in Gangwon-do province formed in Cambrian-Ordovician; and the Jeongseon-gun site, the type locality of C. splendentissima, is a well-known, large limestone areas hosting Trichophorum dioicum J. Jung & H. K. Choi and Scirpus sylvaticus L. var. maximowiczii Regel (Korea National Arboretum, 2012). How to manage the limestone areas are critical to conserving many valuable natural resources, including C. splendentissima. Furthermore, although the limestone preference of the species is understood, evaluations of population size, structures, and rarity have not been conducted. To establish proper, efficient conservation strategy of the species, ecological, phylogenetic, taxonomic features of the species should be understood.
C. splendentissima is distinct with staminate terminal spikes, broad leaves, and glabrous perigynia and grows in limestone areas. Four members of sect. Siderostictae occur in Korea, but a key to the four species is not available. Two versions of keys to four taxa are provided here. Depending on characters available, the keys can be used. The phylogenetic analyses concluded that C. splendentissima, a Korean endemic sedge, is monophyletic and belongs to the Siderostictae clade with the close relationship with C. ciliatomarginata and C. pachygyna. The sister species of C. splendentissima needs to be investigated with a more appropriate dataset comprehensive sampling of the Siderostictae clade. To conserve the endemic and ecological specialist species, further research on genetic and ecological structures at a population level is required.
Key to the four Korean species in Carex sect. Siderostictae
1. Leaf margins ciliolate; perigynia surfaces pubescent ····· ···························································· C. ciliatomarginata
1. Leaf margins not ciliolate; perigynia surfaces glabrous
2. Leaves less than 1 cm in width; perigynia ovate ······ ···································································· C. okamotoi
2. Leaves more than 1 cm in width; perigynia narrowly elliptic or elliptic
3. Leaf abaxial surfaces pubescent; terminal spikes bisexual (androgynous) ·················· C. siderosticta
3. Leaf abaxial surfaces glabrous; terminal spikes unisexual (staminate) ··············· C. splendentissima
Key modified from Park et al. (2016)
1. Terminal spikes bisexual (androgynous); lateral spikes unisexual (staminate) ································ C. siderosticta
1. Terminal spikes unisexual (staminate); lateral spikes bisexual (androgynous)
2. Culms, leaves, perigynia pubescent ····························· ······················································· C. ciliatomarginata
2. Culms, leaves, perigynia glabrous
3. Leaves broad lanceolate or linear lanceolate, 4–8 mm wide; perigynia obovate ········· C. okamotoi
3. Leaves oblanceolate or obovate, 14–46 mm wide; perigynia narrowly elliptic or elliptic ····················· ···················································· C. splendentissima
Acknowledgements
Authors thank Deokgi Nam, a para-taxonomist, for helping with natural habitat surveys of Carex splendentissima and Dr. Seong-Jin Ji at National Science Museum for the species photograph. Herbarium staff members at Korea National Arboretum (KH) and National Institute of Biological Resources (KB) provided assistants for specimen examination. We also thank two anonymous reviewers for their critical comments and suggestions to improve the manuscript.
Notes
Conflict of Interest
The authors declare that there are no conflicts of interest.
References
Appendices
Appendix 1. Voucher specimens of C. splendentissima examined for the distribution map.
KOREA. Gangwon-do: Jeongseon-gun, Jeongseon-eup, 8 Jun 2018, Chung et al. 5159 (KH); Pyeongchang-gun, Daehwa-myeon, Haanmi-ri, 8 May 2012, Nam s.n. (barcode number, NIBRVP0000356733) (KB); Yeongwol-gun, Hanbando-myeon, Ongjeong-ri, 13 May 2013, Kim and Chae s.n. (barcode number, NIBRVP0000612118) (KB).
Appendix 2. GenBank accession numbers for DNA data.
ITS: Carex albursina USA AY757626; C. ciliatomarginata KOR1 JX644818, KOR2 MT811777, CHN AB725706, JPN AB725707; C. gibba JPN DQ998917; C. glossostigma CHN AB725708; C. grandiligulata CHN AB725709; C. kwangsiensis CHN AB725730; C. longshengensis CHN AB725710; C. okamotoi KOR1 AB725711, KOR2 JX644835, KOR3 MT811778; C. omiana JPN DQ998935; C. pachygyna JPN AB725712; C. plantaginea CAN AY757613; C. siderosticta CHN1 AB725713, CHN2 AB725714, JPN1 AB725715, JPN2 AB725716, KOR1 AB725717, KOR2 AB725718, KOR3 MT811783; C. splendentissima KOR1 MT274323, KOR2 MT811781, KOR3 MT811782; C. subcapitata CHN1 AB725719, CHN2 AB725720; C. tsiangii CHN AB725731; C. tumidula JPN AB725721; Scirpus microcarpus AF284961; Trichophorum pumilum AB643647; T. subcapitatum AB679909.
trnL-F: Carex albursina USA AY757554; C. ciliatomarginata KOR1 JX644730, KOR2 MT826862, CHN AB725732, JPN AB725733; C. gibba JPN DQ998970; C. glossostigma CHN AB725734; C. grandiligulata CHN AB725735; C. kwangsiensis CHN AB725756; C. longshengensis CHN AB725736; C. okamotoi KOR1 AB725737, KOR2 JX644729, KOR3 MT826872; C. omiana JPN DQ998988; C. pachygyna JPN AB725738; C. plantaginea CAN AY757540; C. siderosticta CHN1 AB725739, CHN2 AB725740, JPN1 AB725742, JPN2 AB725742, KOR1 AB725743, KOR2 AB725744, KOR3 MT826873; C. splendentissima KOR1 MT316043, KOR2 MT826880, KOR3 MT826882; C. subcapitata CHN1 AB725745, CHN2 AB725746; C. tsiangii CHN AB725757; C. tumidula JPN AB725747; Scirpus microcarpus AF284859; Trichophorum pumilum AB643656; T. subcapitatum AB679914.