Evaluation of the taxonomic rank of the terrestrial orchid Cephalanthera subaphylla based on allozymes
Article information
Abstract
The taxonomic rank of the tiny-leaved terrestrial orchid Cephalanthera subaphylla Miyabe & Kudô has been somewhat controversial, as it has been treated as a species or as an infraspecific taxon, under C. erecta (Thunb.) Blume [C. erecta var. subaphylla (Miyabe & Kudô) Ohwi and C. erecta f. subaphylla (Miyabe & Kudô) M. Hiro]. Allozyme markers, traditionally employed for delimiting species boundaries, are used here to gain information for determining the taxonomic status of C. subaphylla. To do this, we sampled three populations of five taxa (a total of 15 populations) of Cephalanthera native to the Korean Peninsula [C. erecta, C. falcata (Thunb.) Blume, C. longibracteata Blume, C. longifolia (L.) Fritsch, and C. subaphylla]. Among 20 putative loci resolved, three were monomorphic (Dia-2, Pgi-1, and Tpi-1) across the five species. Apart from C. longibracteata, there was no allozyme variation within the remaining four species. Of the 51 alleles harbored by these 17 polymorphic loci, each of the 27 alleles at 14 loci was unique to a single species. Accordingly, we found low average values of Nei’s genetic identities (I) between ten species pairs (from I = 0.250 for C. erecta versus C. longifolia to I = 0.603 for C. falcata vs. C. longibracteata), with C. subaphylla being genetically clearly differentiated from the other species (from I = 0.349 for C. subaphylla vs. C. longifolia to 0.400 for C. subaphylla vs. C. falcata). These results clearly indicate that C. subaphylla is not genetically related to any of the other taxa of Cephalanthera that are native to the Korean Peninsula, including C. erecta. In a principal coordinate analysis (PCoA), C. subaphylla was positioned distant not only from C. falcata, C. longibracteata, and C. longifolia, but also from C. erecta. Finally, K = 5 was the best clustering scheme using a Bayesian approach, with five clusters precisely corresponding to the five taxa. Thus, our allozyme results strongly suggest that C. subaphylla merits the rank of species.
On the Korean Peninsula, the genus Cephalanthera is represented by five taxa (species): Cephalanthera erecta (Thunb.) Blume, C. falcata (Thunb.) Blume, C. longibracteata Blume, and C. longifolia (L.) Fritsch, and C. subaphylla Miyabe and Kudo. The taxonomic rank of the former four species has been well accepted, while the rank of the last has been somewhat controversial. In 1932, Kingo Miyabe and Yushun Kudo described this tiny-leaved, mycoheterotrophic plant in Japan as a new species. In 1953, Jisaburo Ohwi treated it as a variety of C. erecta, C. erecta var. subaphylla (Miyabe and Kudo) Ohwi, and, in 1971, Minosuka Hiroe treated it as a form of C. erecta, C. erecta f. subaphylla (Miyabe and Kudo) M. Hiroe. In recent times, while some authors refer to this taxon as a species (e.g., Lee et al., 2009; Lee, 2011; Sakamoto et al., 2016) others do it as a variety (e.g., Jung et al., 2013; Ministry of Environment, Republic of Korea, 2014). Currently, plant databases do also not consistently name this taxon. For example, Wikispecies (https://species.wikimedia.org/) and World Checklist of Selected Plant Families (WCSP, https://wcsp.science.kew.org/) treat this taxon (whatever it is at species, varietal or formal ranks) under the synonymy of C. erecta; in contrast, The Plant List (https://www.theplantlist.org/) and Tropicos (http://www.tropicos.org/) use C. erecta f. subaphylla as an accepted name. Accordingly, it is necessary to evaluate the taxonomic rank of this taxon using appropriate, independent taxonomic tools.
The nearly neutral allozyme markers have been useful for identifying species boundaries in a variety of plant groups (Crawford, 1989; Arduino et al., 1996; Harris and Abbott, 1997; Chung et al. 2005; Hedrén and Nordström, 2009; López-Pujol et al. 2012; Chung et al., 2018). A review on orchid allozyme-based genetic diversity studies revealed that most orchid species examined so far have ‘diagnostic’ or unique alleles for species delimitation at several isozyme loci (reviewed in Chung and Chung, 2012). The more recent study of Chung et al. (2018), focused on three Cephalanthera species in South Korea (C. erecta [three populations], C. falcata [five populations], and C. longibracteata [three populations]), revealed no allozyme variation within any of the species using 20 putative loci; all populations of each species were fixed for one allele at all loci (expected heterozygosity at population level and sample as a whole, HeP and HeS = 0). However, there was considerable genetic variation between species, with 13 loci being useful for the three Cephalanthera species delimitation. Among the 30 alleles harbored by these 13 loci, each of the three alleles at four loci were unique (‘diagnostic’) to a single species (Chung et al., 2018). The presence of these species-specific alleles was the reason for the relatively low Nei’s (1978) genetic identities found between species pairs: 0.400 for Cephalanthera erecta vs. C. falcata; 0.500 for C. erecta vs. C. longibracteata; and 0.600 for C. falcata vs. C. longibracteata (Chung et al., 2018). Chung et al. (2018) also found no hybrids in sympatric populations at the landscape level in Yeonwhasan Provincial Park (YPP, 600 × 600-m area [36 ha]; altitude, 230 m asl). Thus, it seems relatively easy or simple to use alleles at several loci to identify species.
In this study, we have conducted an allozyme study of three populations (a total of 15 populations) of each of five Cephalanthera taxa native to the Korean Peninsula (C. erecta, C. falcata, C. longibracteata, C. longifolia, and C. subaphylla) to gain information for determining the taxonomic rank of C. subphylla. Is this taxon differentiated genetically from C. erecta and from other congeneric species? Does it, thus, merit the species rank? The previous study (Chung et al., 2018) revealed that C. erecta, C. falcata, C. longibracteata, and C. subaphylla in southern Korea lack allozyme variation within each species. Although not being the main objective of this study, we have also evaluated the levels of genetic variation in C. longifolia in southern Korea.
Materials and Methods
Population sampling
To survey allozyme variation and to screen unique (diagnostic) alleles to each species, we collected leaf samples from three populations representing the five Cephalanthera species across South Korea (Fig. 1). A total of 360 samples were collected from 15 populations: ERE-1 (n = 17), ERE-2 (n = 18), and ERE-3 (n = 20) of C. erecta; FAL-1 (n = 31), FAL-2 (n = 19), and FAL-3 (n = 39) of C. falcata; FOL-1 (n = 31), FOL-2 (n = 10), and FOL-3 (n = 15) of C. longifolia; LON-1 (n = 90), LON-2 (n = 21), and LON-3 (n = 13) of C. longibracteata; and SUB-1 (n = 14), SUB-2 (n = 10), and SUB-3 (n = 12) of C. subaphylla (Fig. 1, Table 1). Data for ERE-2, ERE-3, FAL-1 FAL-2, LON-1, and LON-3 (as ERE, YPP-3 of C. erecta, FAL, YPP-3 of C. falcata, LON, and YPP- 3 of C. longibracteata, respectively) were taken from the previous study (Fig. 1 and Table 2 in Chung et al., 2018). To minimize the damage to these plants, we cut just 1 cm from the tip of one shoot.
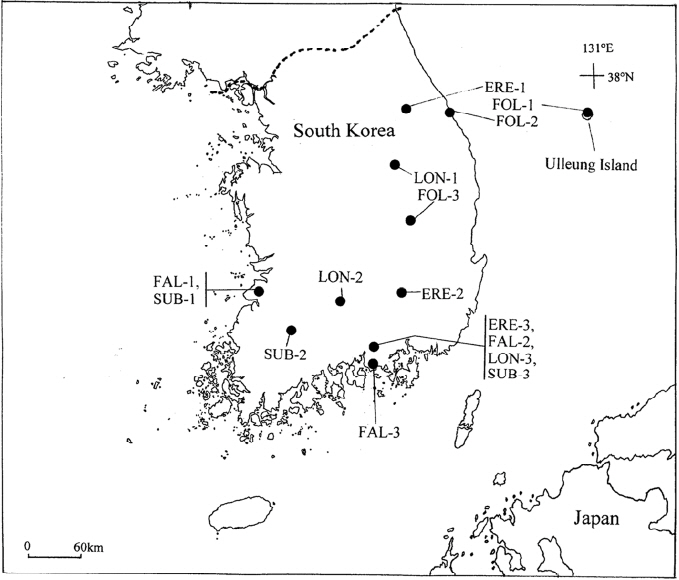
Locations of sampled populations of the five Cephalanthera species in South Korea: C. erecta, ERE-1 (Jeongsun County), ERE-2 (Changryeong County), and ERE-3 (Yeonwhasan Provincial Park, YPP); C. falcata, FAL-1 (Buan County), FAL-2 (YPP), and FAL-3 (Gosung County); C. longifolia, FOL-1 (Ulleung Island), FOL-2 (Samchuk City), and FOL-3 (Uisung County); C. longibracteata, LON-1 (Danyang County), LON-2 (Hamyang County), and LON-3 (YPP); C. subaphylla, SUB-1 (Buan County), SUB-2 (Damyang County), and SUB-3 (YPP).
Isozyme electrophoresis
We transported leaf samples on an ice box after collection and kept them in a refrigerator once at the M.G.C. laboratory. Within one day we cut and crushed them with a precooled mortar and pestle in a phosphate polyvinylpyrrolidone extraction buffer (Mitton et al., 1979). We absorbed enzyme extracts onto 4 × 6-mm wicks cut from Whatman 3 MM chromatography paper (Whatman International, Maidstone, UK), which were then stored at −70°C until needed. We determined allozyme variation via horizontal starch-gel electrophoresis. We prepared 13% gels about 12 h before gel running. We used a Poulik system (Poulik, 1957) to resolve alcohol dehydrogenase (Adh; dimer, E.C. 1.1.1.1), cathodal peroxidase (Cpx; monomer, E.C. 1.11.1.7), diaphorase (Dia-1, Dia-2; monomer, E.C. 1.6.99.-), fluorescent esterase (Fe-1, Fe-2; monomer, E.C. 3.1.1.-), leucine aminopeptidase (Lap; monomer, E.C. 3.4.11.1), malic enzyme (Me; dimer, E.C. 1.1.1.40), phosphoglucoisomerase (Pgi-1, Pgi-2; dimer, E.C. 5.3.1.9), phosphoglucomutase (Pgm-1, Pgm-2; monomer, E.C. 5.4.2.2), and triosephosphate isomerase (Tpi-1, Tpi-2; dimer, E.C. 5.3.1.1). We also used the morpholine citrate buffer system (pH 6.1) of Clayton and Tretiak (1972) to resolve isocitrate dehydrogenase (Idh; dimer, E.C. 1.1.1.42), malate dehydrogenase (Mdh-1, Mdh-2; dimer, E.C. 1.1.1.37), 6-phosphogluconate dehydrogenase (6Pgd-2; dimer, E.C. 1.1.1.44), and shikimic acid dehydrogenase (Skdh-1, Skdh-2; monomer, E.C. 1.1.1.25). Following Soltis et al. (1983) stain recipes (except for diaphorase; Cheliak and Pitel, 1984), we stained starch gels for the 13 enzyme systems, which produced 20 putative loci. We designated putative loci sequentially, with the most anodally migrating isozyme designated as ‘1,’ the next ‘2,’ and so on. We also designated different alleles within each locus sequentially by alphabetical order (‘a’, ‘b’, ‘c’, ‘d’, ‘e’). The locus 6Pgd-1 was variable among species but inconsistently stained; thus, we excluded this locus from data analysis.
Data analysis
We considered a locus to be polymorphic when two or more alleles were observed, regardless of their frequencies. We calculated allele frequencies for all populations to determine species’ diagnostic alleles using the program FSTAT (Goudet, 1995). We further estimated the genetic diversity parameters within populations using the programs POPGENE (Yeh et al., 1999) and FSTAT: percentage of polymorphic loci (%PP), mean number of alleles per locus (AP), and Hardy-Weinberg (H-W) expected heterozygosity or Nei’s (1978) gene diversity (HeP). In addition, these parameters were also estimated for the total samples as a whole (i.e., the species level; %PS, AS, and HeS). With the raw genotypic data, we also estimated genetic divergence among populations/species by calculating Nei’s (1978) unbiased genetic identity (I) for all pair of populations using POPGENE.
As complementary analysis to access genetic divergence among the studied taxa, we performed a principal coordinate analysis (PCoA) with GenAlEx 6.5 (Peakall and Smouse, 2012) based on codominant genotypic distances. In addition, we used Structure 2.3.4 (Pritchard et al., 2000) to analyze the allozyme data with a Bayesian approach. Posterior probabilities of the data [ln Pr(X|K)] for each K were obtained for K = 1 to K = 15 clusters using a model assuming non-admixture and uncorrelated allele frequencies. Ten runs were completed for each K, with a Markov Chain Monte Carlo (MCMC) of 500,000 iterations, following a burn-in period of 50,000 iterations. We inferred the most likely value of K by the ΔK statistic of Evanno et al. (2005), with the aid of Structure Harvester (Earl and vonHoldt, 2012). Since the ΔK method tends to identify K = 2 as the top level of hierarchical structure (Janes et al., 2017), we combined it with the method of selecting the smallest K after ln Pr(X|K) values reached a plateau (Pritchard et al., 2010).
Results
Allele profiles among the five Cephalanthera species
Among 20 putative loci resolved, three were monomorphic (Dia-2, Pgi-1, and Tpi-1) across the five species. Of the 51 alleles harbored by the remaining 17 loci, each of the 27 alleles at 14 loci were unique to a single species, and were thus useful for Cephalanthera species delimitation (Table 1). Nine alleles (Adhb, Fe-1c, Fe-2b, Lapa, Mdh-1a, Mdh-2d, 6Pgd-2b, Pgi-2a, and Pgm-2c) were unique to C. erecta, one allele (6Pgd-2d) to C. falcata, five alleles (Fe-2d, Idhc, Mdh-2c, Meb, and Skdh-1c) to C. longifolia, five alleles (6Pgd-2c, Pgi-2b, Pgm-1a, Pgm-2a, and Pgm-2b) to C. longibracteata, and seven alleles (Fe-2a, Idhb, Mdh-2b, Pgi-2c, Pgm-2d, Skdh-2b, and Tpi-2a) to C. subaphylla (Table 1). On the other hand, C. erecta and C. longibracteata shared one allele (Skdh-1b), C. erecta, C. falcata, and C. longibracteata one (Idha), C. erecta, C. falcata, and C. subaphylla one (Dia-1a), C. erecta, C. falcata, C. longibracteata, and C. subaphylla one (Mea), C. erecta, C. falcata, C. longifolia, and C. longibracteata two (Skdh-2a and Tpi-2b), C. erecta, C. longibracteata, and C. subaphylla two (Cpxb and Pgm-1b), C. falcata and C. longifolia four (Cpxa, Pgi-2d, Pgm-1c, and Pgm-2e), C. falcata and C. longibracteata four (Fe-1b, Fe-2c, Mdh-1b, and Mdh-2a), C. falcata and C. subaphylla two (Lapc and Skdh-1a), C. falcata, C. longifolia, C. longibracteata, and C. subaphylla one (Adha), C. longifolia and C. longibracteata two (Dia-1b and Lapb), and C. longifolia and C. subaphylla three (Fe-1a, Mdh-1c, and 6Pgd-2a) (Table 2).
As representative zymograms (in color photos) for allelic variation patterns of the five Cephalanthera species were clearly presented in the previous study (Fig. 2 in Chung et al., 2018), it may not necessary to show them repeatedly in this study.
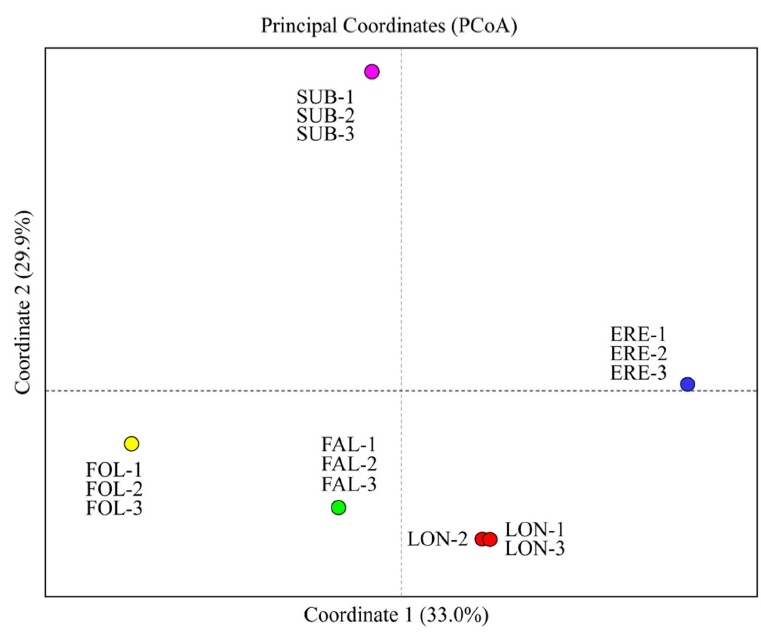
The principal coordinate analysis (PCoA) of the 15 studied populations of the five Cephalanthera species in South Korea. Blue circles, populations of C. erecta (ERE-1 to ERE-3); green circles, populations of C. falcata (FAL-1 to FAL-3); yellow circles, populations of C. longifolia (FOL-1 to FOL-3); red circles, populations of C. longibracteata (LON-1 to LON-3); pink circles, populations of C. subaphylla (SUB-1 to SUB-3).
Nei’s genetic identity, PCoA, and Bayesian clustering approach
We found that C. subaphylla was genetically differentiated clearly from the other Cephalanthera species examined, including C. erecta (from I = 0.349 for C. subaphylla vs. C. longibracteata to 0.400 for C. subaphylla vs. C. falcata) (Table 2). Values of Nei’s genetic identity (I) between ten Cephalanthera species pairs were in general low (from I = 0.250 for C. erecta vs. C. longifolia to 0.603 for C. falcata vs. C. longibracteata) (Table 2).
PCoA results were in good agreement with Nei’s I values (Fig. 2). The first two components accounted for 62.9% (axis 1 = 33.0%; axis 2 = 29.9%) of the total genetic variance (Fig. 2). When the 15 populations were plotted along the first two axes, the five taxa were genetically clearly separated (Fig. 2). The genetic distinctiveness of the five Korean Cephalanthera taxa was also well supported by the STRUCTURE results, given that K = 5 was the best clustering scheme according to both the “plateau” method and the ΔK statistic (Fig. 3), and these five clusters exactly corresponded to the five taxa.
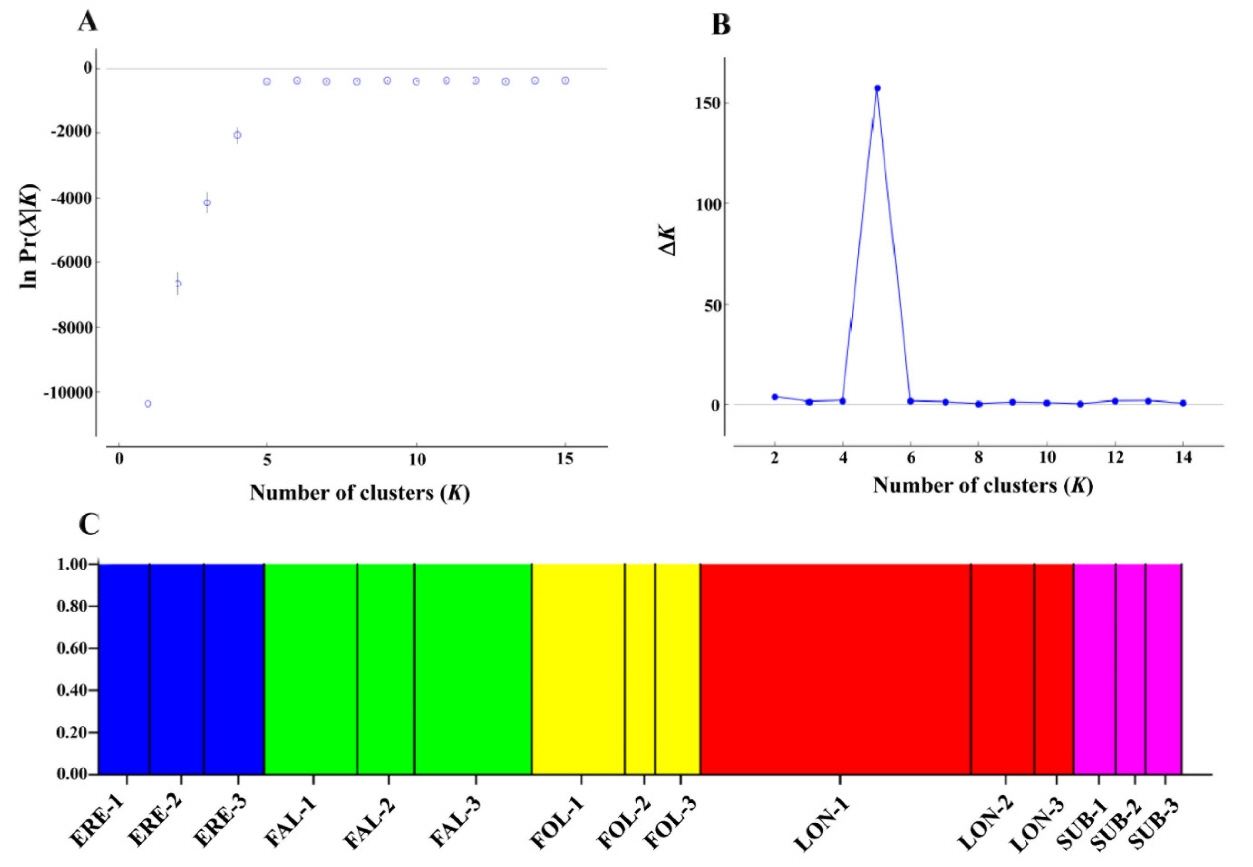
Results of STRUCTURE analysis for all studied individuals of the five Cephalanthera species in South Korea. The most likely K was estimated by choosing the smallest K after the log probability of data [ln Pr(X|K)] values reached a plateau (Pritchard et al., 2010) (A), and the ΔK statistics (Evanno et al., 2005) (B), using Structure Harvester (Earl and vonHoldt, 2012), and Bayesian clustering analysis based on the allozyme data for 15 populations when K = 5 (C). Blue cluster, populations of C. erecta (ERE-1 to ERE-3); green cluster, populations of C. falcata (FAL-1 to FAL-3); yellow cluster, populations of C. longifolia (FOL-1 to FOL-3); red cluster, populations of C. longibracteata (LON-1 to LON-3); pink cluster, populations of C. subaphylla (SUB-1 to SUB-3).
Genetic diversity
Except for C. longibracteata (two alleles were found at Pgm-1 and Pgm-2 in LON-2) (Table 1), there was no allozyme variation within the Cephalanthera species in southern Korea (expected heterozygosity, HeP and HeS = 0) and all populations of each species were fixed for one allele at each locus. However, C. longibracteata also harbored extremely low levels of genetic diversity at both population and species levels (%PS = 10%, AS = 1.10, HeS = 0.013; %PP = 3.3%, AP = 1.03, HeP = 0.011).
Discussion
Evaluation of the taxonomic rank of C. subaphylla
We found a total of 27 unique alleles for the five Cephalanthera species; nine alleles to C. erecta, one to C. falcata, four to C. longifolia, six alleles to C. longibracteata, and seven to C. subaphylla. These allele patterns among species strongly suggest that allozymes could be useful to solve uncertainty associated with species/taxa boundaries of terrestrial orchid taxa.
Chung and Chung (2012) conducted a meta-analysis of allozyme-based Nei’s genetic identity (I) on Orchidaceae and found a mean of 0.955 for conspecific populations from 84 studies (I values ranged from 0.756 to 1.000 ) and a mean of 0.453 for congeneric orchid species pairs from 190 studies (range = 0.000–0.978). Nei’s I estimates calculated among the five taxa of Cephalanthera studied herein (mean = 0.410; range = 0.250–0.603) are, as one might expect, close to those reported for congeneric orchids (mean I = 0.453; Chung and Chung, 2012). In particular, we found that I values between C. subaphylla vs. other four species are even lower (ranged from 0.349 to 0.400) than the expected ones (mean I = 0.453) (Chung and Chung, 2012). These results clearly indicate that C. subaphylla is not genetically related to any of the other taxa of Cephalanthera that are native to the Korean Peninsula, including C. erecta. The PCoA results also suggest the separation of C. erecta and C. subaphylla as distinct species; C. subaphylla is placed well far away not only from C. falcata, C. longibracteata, and C. longifolia, but also from C. erecta. The STRUCTURE results are also supporting the treatment of C. subaphylla as a taxonomic entity clearly differentiated from C. erecta.
Independent from this study, Lee (2011) noted in her description of C. subaphylla native to southern Korea that “based on a phylogenetic analysis, it seems reasonable to treat this taxon as a species rather than a ‘form’ under C. erecta”. In addition, Yuki Sakamoto and collaborators conducted a molecular phylogenetic analysis on Cephalanthera and found that C. subaphylla is a sister species of C. erecta based on materials from Japan (Y. Sakamoto et al., unpublished data); thus the former can be recognized as an independent species (this information is shown in Sakamoto et al., 2016). In essence, our allozyme data as well as the unpublished molecular phylogenetic analyses strongly suggest that C. subaphylla merits species rank. The present study confirms the usefulness of allozymes as a complementary approach of molecular phylogenetic analysis to delimit species boundaries of orchids.
Genetic diversity
The five Cephalanthera species native to South Korea are genetically depauperate compared with C. longifolia in central Italy (%PS = 55.6%, AS = 1.67, HeS = 0.188; %PP = 48.1%, AP = 1.59, HeP = 0.168) (Scacchi et al., 1991) and with C. rubra in central Italy (%PS = 66.7%, AS = 1.67, HeS = 0.180; %PP = 33.3%, AP = 1.33, HeP = 0.127) (Scacchi et al., 1991) and in northeast Poland (%PS = 53.9%, AS = 1.54, HeS = 0.125; %PP = 13.9%, AP = 1.14, HeP = 0.059) (Brzosko and Wroblewska, 2013). Chung et al. (2018) suggest that historical factors such as the Quaternary climate oscillations played a major role in shaping current levels of genetic diversity in the Cephalanthera species native to the Korean Peninsula. The Korean populations of C. erecta (a warm-temperate/temperate element) and C. falcata (a warm-temperate element) would have been established by a single introduction from a genetically depauperate ancestral population, likely located outside the Korean Peninsula. On the other hand, since C. longifolia, C. longibracteata, and C. subaphylla are boreal/temperate elements, they may have survived the Last Glacial Maximum in microrefugia located in low elevation regions within the Peninsula; in these microrefugia, they would have been subjected to population bottlenecks reducing their genetic diversity. In particular, C. longifolia and C. subaphylla show a complete lack of allozyme diversity at 20 loci, which may be primarily attributed to random genetic drift for a long period of time, a scenario that is compatible with the rarity of these two species within the Korean Peninsula.
Acknowledgements
The authors thank Beom Jin Shim and Myeong Soon Park for laboratory assistance. This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2013R1A1A2063524 to MYC and NRF-2013R1A1A3010892 and NRF-2017R1A2B4012215 to MGC) and was carried out as part of the Infrastructure for the Conservation and Restoration of Rare and Endemic Plants in Korea National Arboretum that supported to MGC from 2015 to 2019.
Notes
Conflict of Interest
The authors declare that there are no conflicts of interest.


