New polymorphic microsatellite markers for the endangered fern Ceratopteris thalictroides (Parkeriaceae)
Article information
Abstract
Ceratopteris thalictroides is a semi-aquatic fern with a circumtropical distribution. Because this species is designated internationally on the IUCN Red List as requiring at least some concern, Korean populations are of great concern for the species’ long-term survival, as they are at the northern limit of the species distribution. To establish an effective conservation strategy for those populations at the genetic level, we used the Mi-Seq platform to develop three sets of 25 polymorphic microsatellite markers for C. thalictroides, which is endangered in Korea. In populations sampled from Busan and Gochang, the number of alleles ranged from 2 to 13 (average of 5.64), and plants presented an expected heterozygosity of 0.000 to 0.860. These markers will be useful for evaluating the genetic status and conserving Korean populations of C. thalictroides more effectively.
Ceratopteris thalictroides (L.) Brongn. (Parkeriaceae) is a semi-aquatic fern with circum-tropical distribution, mainly growing in paddy fields, ponds, or marshes (Watano and Masuyama, 1994). This genus contains four species (Lloyd, 1974). Whereas three of those species are diploid plants, C. thalictroides is tetraploid (Hickok, 1977; Masuyama and Watano, 2005). Although C. thalictroides is widely distributed throughout tropical and subtropical regions, it is internationally considered to be of as least some concern, based on the IUCN Red List of Threatened Species.
In Korea, this species was first recorded at Suncheon and Gwangyang, in the southern regions of the Peninsula. Since then, more populations have been identified. This species is now designated as endangered in Korea and has been assigned legal protection for eight extant populations (Seocheon, Gunsan, Jeongeup, Buan, Iksan, Gochang, Gwangju, and Busan). These Korean populations are clearly critical because they represent the northern edge of the distribution range, possibly providing potential for further expansion of the species if those plants are able to adapt to selection pressures from such marginal environments (Kawecki, 2008). Therefore, efforts to conserve Korean populations are needed if we are to achieve long-term survival for C. thalictroides.
In China, researchers are also taking various steps to conserve the populations of C. thalictroides. There, this species has been designated as endangered in an effort to protect and manage its native habitats (Yu, 1999). In addition, its genetic variability has been analyzed by using various markers such as random amplified polymorphic DNA, inter-simple sequence repeat, and microsatellites (Dong et al., 2008; Yang et al., 2016). Although 30 microsatellite markers have been established for Chinese populations of C. thalictroides (Yang et al., 2016), they are not adequate for frequently examining genetic polymorphism. Furthermore, it is cost-effective for such markers to be organized as “sets” if researchers are to continue periodic genetic monitoring. Therefore, we have developed new polymorphic microsatellite sets for evaluating the genetic status of Korean populations of C. thalictroides and conserving those plants.
Materials and Methods
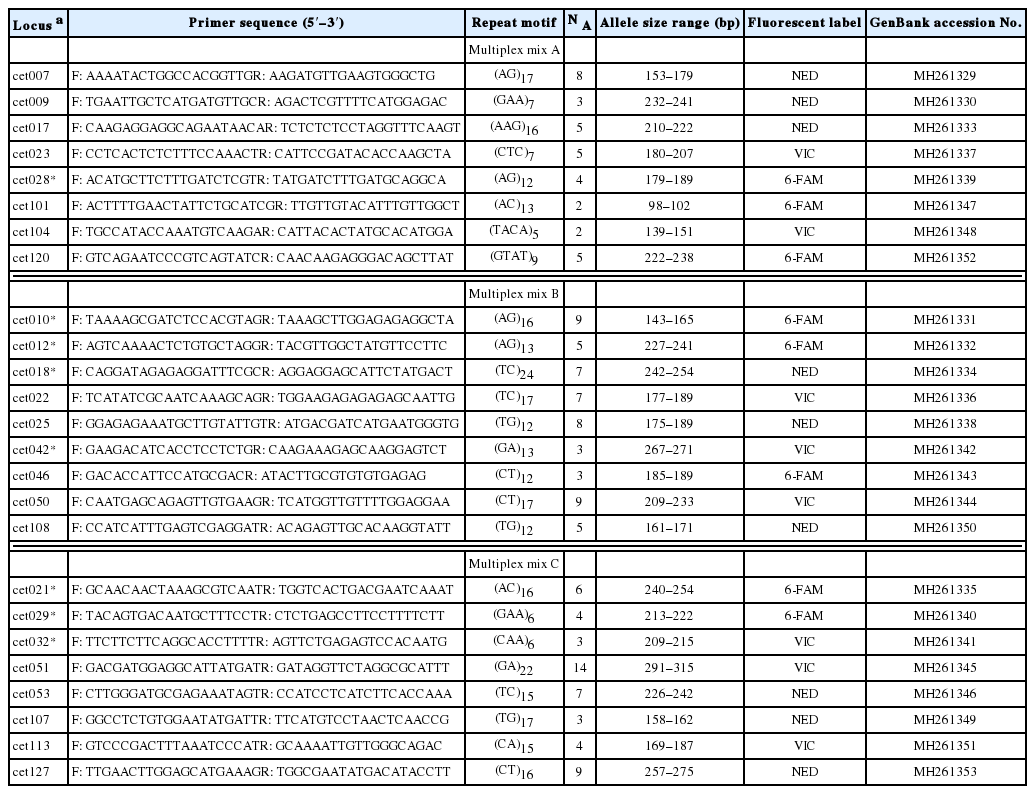
For these new microsatellite markers, total genomic DNA was obtained from a fresh leaf collected from an individual plant of C. thalictroides growing at the Dongnim reservoir, Gochang, Korea. The extraction procedure involved an MG Plant Genomic DNA Extraction SV Miniprep Kit used according to the manufacturer’s protocol (Macrogene Inc., Seongnam, Korea). After the quality of the genomic DNA was checked by gel electrolysis, we generated a shotgun library by using the Illumina Mi-Seq platform (LAS, Seoul, Korea). In all, 5,256,220 paired-ends with read lengths of approximately 300 bp were obtained. Based on screening with the SSR_Pipeline v.0951 infrastructure (Miller et al., 2013), we identified a 22,990 microsatellite motif with flanking regions larger than 100 bp that contained di-, tri-, or tetra-nucleotide repeats that were at least 12, 6, or 5 times, respectively, in size. Following the method of Cho et al. (2015), we assembled groupings of 50 to 100 reads that were reference guide-mapped on the whole raw reads, using the Geneious R10.1.3 software platform (Kearse et al., 2012). From this, we selected reads that had two separate alleles and no additional single nucleotide polymorphisms in the flanking region. After eliminating any duplication through de novo assembly, we used the final chosen reads to develop microsatellite markers. We then utilized the Primer 3 software in Geneious program R10.1.3 to design 81 primer pairs (18–22 bp long), using a melting temperature of 53–60°C and GC contents of 35–65%. The forward primers added three sets of M13 tag sequences (5′-CACGACGTTGTAAACGAC-3′, 5′-TGTGGAATT GTGAGCGG-3′, and 5′-CTATAGGGCACGCGTGGT-3′) with 6-FAM, VIC, and NED fluorescent dye, respectively. For multiplex polymerase chain reaction (PCR), each primer set comprised nine primers with lengths of 100–300 bp.
To test the effectiveness of these new microsatellite loci, we sampled 63 individuals of C. thalictroides from two populations (Dongnim reservoir, Gochang, n = 33; Is. Doonchido, Busan, n = 30). For each locus, PCR amplification was performed in a final volume of 5 μL that contained 15 to 20 ng of extracted DNA, 2.5 μL of 2× Multiplex PCR master mix (Qiagen, Valencia, CA, USA), 0.01 μM for the forward primer, 0.2 μM for the reverse primer, and 0.1 μM for the M13 primer. The PCR protocol included an initial denaturation for 15 min at 95°C; followed by 30 cycles, each consisting of denaturing for 30 s at 94°C, 1.5 min of annealing at 52°C, and extension for 1.5 min at 72°C; with a final extension for 10 min at 72°C. The PCR products were analyzed using an ABI 3730XL sequencer with GeneScan-500LIZ Size Standard (Applied Biosystems, Foster City, CA, USA). Allele sizes and the peaks of each sample were determined via Peak Scanner software 2 (Applied Biosystems). In analyzing the tetraploid microsatellite data, we were not able to make exact determinations of allele frequencies and patterns due to the uncertainty of multiple alleles (Dufresne et al., 2014). Therefore, we calculated the number of alleles (N A), the Shannon-Wiener Diversity Index (H′), and the expected heterozygosity (H E) using Atetra 1.2 software (van Puyvelde et al., 2010), which was developed for analyzing codominant microsatellite data in tetraploid species.
Results and Discussion
We have produced useful sets of microsatellite markers for analyzing genetic diversity and establishing conservation strategies for Ceratopteris thalictroides, an endangered plant. Of the 81 primer pairs studied here, 25 microsatellite markers were polymorphic, and 17 were found to be monomorphic in two of the examined populations (Table 1). All developed markers have been deposited in GenBank (Table 1). Among the 25 polymorphic microsatellite loci, the number of alleles per locus ranged from 2 to 13, with an average of 5.64. Values for (H E) were 0.000 to 0.860, while H′ ranged from 0.000 to 2.152 (Table 2). These results are similar to those reported from a previous study in China (Yang et al., 2016) that involved RAD tag sequencing (N A, 4–10; H E, 0.264–0.852). However, because the Korean Peninsula is a northern limit for the distribution of C. thalictroides, our findings suggest that the marker sets described here include considerable polymorphism.
Acknowledgements
This work was supported by the National Institute of Biological Resources of Korea (Grant No. NIBR201703101).
Notes
Conflict of Interest
Authors declare that there is no conflict of interest.