Molecular systematics of Poaceae based on eight chloroplast markers, emphasizing the phylogenetic positions of Korean taxa
Article information
Abstract
This study was conducted to clarify the phylogenetic position and relationships of Korean Poaceae taxa. A total of 438 taxa including 155 accessions of Korean Poaceae (representing 92% and 72% of Korean Poaceous genera and species, respectively) were employed for phylogeny reconstruction. Sequence data of eight chloroplast DNA markers were used for molecular phylogenetic analyses. The resulted phylogeny was mostly concordant with previous phylogenetic hypotheses, especially in terms of subfamilial and tribal relationships. Several taxa-specific indels were detected in the molecular phylogeny, including a 45 bp deletion in rps3 (PACMAD [Panicoideae, Arundinoideae, Chloridoideae, Micrairoideae, Aristidoideae, Danthonioideae] clade), a 15 bp deletion in ndhF (Oryzeae + Phyllorachideae), a 6 bp deletion in trnL-F (Poeae s.l.), and two (17 bp and 378 bp) deletions in atpF-H (Pooideae). The Korean Poaceae members were classified into 23 tribes, representing eight subfamilies. The subfamilial and tribal classifications of the Korean taxa were generally congruent with a recently published system, whereas some subtribes and genera were found to be non-monophyletic. The taxa included in the PACMAD clade (especially Andropogoneae) showed very weak and uncertain phylogenetic relationships, presumably to be due to evolutionary radiation and polyploidization. The reconstructed phylogeny can be utilized to update the taxonomic positions of the newly examined grass accessions.
INTRODUCTION
Poaceae (nom. alt. Gramineae) are the fifth largest family of angiosperms and the second largest family of monocots, containing about 700 genera and 11,000 to 12,000 species (Clayton and Renvoize, 1986; Watson and Dallwitz, 1992; Kellogg, 2015; Soreng et al., 2017). The members of Poaceae are distributed on all continents and are estimated to account for approximately 20% of the earth's vegetation (Shantz, 1954). They are also important food resources for mankind. For example, rice (Oryza sativa L.), wheat (Triticum aestivum L.), and maize (Zea mays L.) are vital food resources that account for more than half of the calories consumed by humans (Raven and Johnson, 1995).
Due to the tremendous ecological and economic importance of Poaceae, numerous taxonomic and phylogenetic studies focusing on this species-rich family have been conducted (Brown, 1810; Palisot de Beauvois, 1812; Kunth, 1833; Tateoka, 1957; Prat, 1960; Stebbins and Crampton, 1961; Jacques-Félix, 1962; Hilu and Wright, 1982; Caro, 1982; Clayton and Renvoize, 1986; Kellogg and Campbell, 1987; Hamby and Zimmer, 1988; Doebley et al., 1990; Watson and Dallwitz, 1992; Cummings et al., 1994; Nadot et al., 1994; Barker et al., 1995; Clark et al., 1995; Soreng and Davis, 1998; Clark et al., 2000; Grass Phylogeny Working Group, 2001; Duvall et al., 2007; Simon, 2007; Bouchenak-Khelladi et al., 2008; Saarela and Graham, 2010; Grass Phylogeny Working Group II, 2011; Kellogg, 2015; Soreng et al., 2017; Saarela et al., 2018; Soreng et al., 2022). Although these studies have led to major changes thus far, the classification system of Poaceae has gradually been stabilizing. Currently, lineages of Poaceae are classified into 12 subfamilies, including three basal subfamilies (Anomochlooideae, Pharoideae, Puelioideae) and two major clades, namely the BOP (Bambusoideae, Oryzoideae, Pooideae) and PACMAD (Panicoideae, Arundinoideae, Chloridoideae, Micrairoideae, Aristidoideae, Danthonioideae) clades (Soreng et al., 2017; Saarela et al., 2018). Attempts have also been made to provide stronger evidence of the phylogeny of certain grass lineages that were not fully resolved in previous studies using plastomes or nuclear genes (Wu and Ge, 2012; Fisher et al., 2016; Dunning et al., 2019; Huang et al., 2022).
Among Korean flora, Poaceae are the third largest family, comprising 212 species representing six subfamilies, 21 tribes, and 94 genera (Lee, 2007). Relatively few taxonomic studies, however, have focused on Korean grasses, although there have been some morphological examinations of well-known genera, such as Miscanthus Andersson (Lee, 1964a, 1964b, 1964c, 1964d) and Poa L. (Jung and Chung, 2008). There have also been molecular phylogenetic studies of common genera, including Echinochloa P. Beauv. (Lee et al., 2014) and Zoysia Willd. (Cheon et al., 2021; Oh et al., 2021), but with limited taxon sampling. Thus far, researchers have disregarded the overall phylogenetic relationships and classifications of Korean Poaceaeous taxa. Therefore, it is necessary to review the current classification system based on detailed phylogenetic information of each plant species for the efficient management and conservation of Korean Poaceous taxa.
In this paper, we present the molecular phylogeny of Poaceae based on the sequences of eight chloroplast markers (atpF-H, psbK-I, trnH-psbA, trnL-F, matK, rbcL, ndhF, rps3), emphasizing the phylogenetic relationships or positions of Korean taxa within the family. We address taxonomic issues pertaining to phylogenetically problematic poaceaous groups for further comprehensive research.
MATERIALS AND METHODS
Taxon sampling
A total of 438 taxa (species or infraspecies) representing 12 subfamilies, 52 tribes, and 297 genera of Poaceae were included in the phylogenetic study. Of these, 155 Korean grass taxa representing 87 genera were newly sampled for the present study (Appendix 1). These correspond to 92% (at the genus level) and 72% (at the species level or below) of all known Poaceae taxa in Korea (Lee, 2007). Species lacking collecting records, rare species, or those only distributed in North Korea were not included in the present study. The names of subfamilies, tribes, and subtribes accepted in the phylogenetic classification system of the Poaceae (Soreng et al., 2017, 2022) were employed throughout the study. The acceptance of scientific names of several controversial taxa was based on The Plant List v1.1 (2013) (http://www.theplantlist.org), Tropicos v3.3.2 (2022) (http://www.tropicos.org), and Plants of the World Online (2022) (http://www.plantsoftheworldonline.org).
Most of the samples for DNA extraction were collected from the field. Fresh young leaves or leaves dried with silica gel were used for DNA extraction. The DNA of some species was obtained from dried specimens deposited at the herbarium of the National Arboretum (KH) and the herbarium of Hallym University (HHU). Voucher specimens of the plants used for DNA extraction were deposited at the HHU and the herbarium of the National Institute of Biological Resources of Korea (KB) (Appendix 1).
Eight plastid genome markers including four non-coding regions (atpF-H, psbK-I, trnH-psbA, and trnL-F) and four coding regions (matK, ndhF, rbcL, and rps3) were employed for the molecular phylogeny reconstruction (Fig. 1). A total of 154 atpF-H sequences, 130 psbK-I sequences, 152 trnH-psbA sequences, 130 trnL-F sequences, 129 matK sequences, 115 ndhF sequences, 131 rbcL sequences, 143 rps3 sequences were newly obtained for the Korean Poaceae taxa (Appendix 1). Various number of sequence data (85 atpF-H, 85 psbK-I, 98 trnH-psbA, 192 trnL-F, 281 matK, 274 ndhF, 270 rbcL, and 108 rps3; see Appendix 1) were downloaded from the GenBank and combined with the newly obtained sequences. Altogether, variable number of nucleotide sequences (239 atpF-H, 215 psbK-I, 249 trnH-psbA, 316 trnL-F, 403 matK, 385 ndhF, 394 rbcL, and 247 rps3) were included for the phylogenetic analyses (Table 1). Seven taxa in neighboring families, Joinvilleaceae and Ecdeiocoleaceae were selected as outgroups in the phylogenetic analyses (Stevens, 2017).
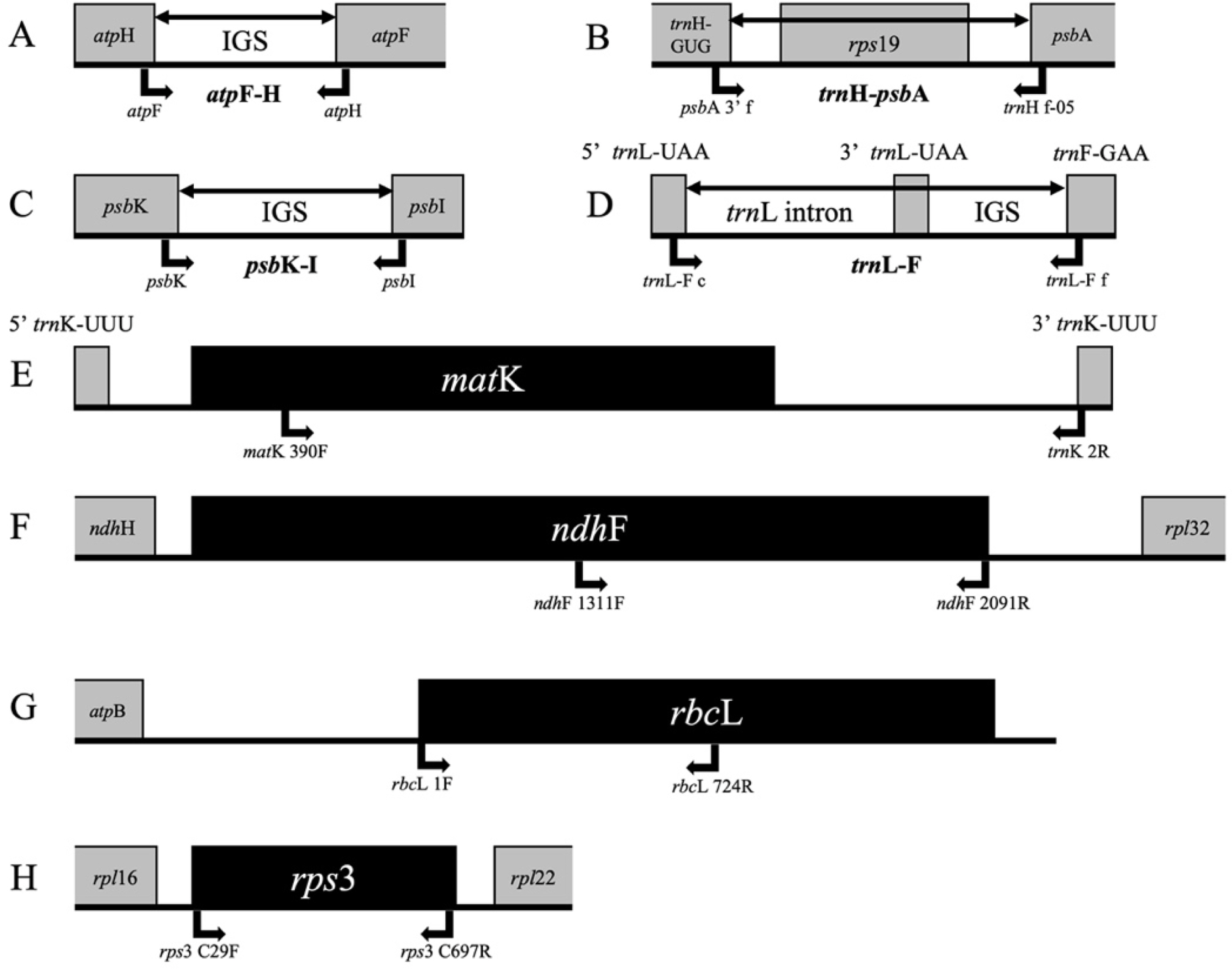
Structure of eight plastid regions examined for the present study. Arrows indicate the forward and reverse primers employed for polymerase chain reaction and sequencing. A. atpF-H (Lahaye et al., 2008). B. trnH-psbA. C. psbK-I (Lahaye et al., 2008). D. trnL-F (Taberlet et al., 1991). E. matK (Cuénoud et al., 2002; Johnson and Soltis., 1995). F. ndhF (Romaschenko et al., 2010). G. rbcL (Fay et al., 1997; Olmstead et al., 1992). H. rps3 (Peterson et al., 2010).
DNA extraction and polymerase chain reaction
DNA was extracted from fresh leaves using CTAB (Doyle and Doyle, 1987) or DNeasy plant mini-kits (Qiagen Inc., Hilden, Germany) following the manufacturer's instructions in each case. The extracted DNA was electrophoresed on 1% agarose gel, stained with ethidium bromide, and compared with the markers on the UV trans-illuminator for DNA quality and for a concentration check. Polymerase chain reaction (PCR) was performed with a final volume of 50 μL, and the concentration of the PCR reaction mixture was 5 units/μL TaKaRa Ex Taq, 20 mM Mg2+ plus 10× Ex Taq Buffer, 2.5 mM dNTP Mixture (TaKaRa Biotechnology Co., Dalian, China), template DNA 20–50 ng, 5 pmol-specific primer pair (Fig. 1), and distilled water. The PCR cycling conditions were pre-denaturation at 95°C for 5 min, followed by 94–95°C denaturation for 40–60 s, annealing for 52–57°C for 40–60 s and 72°C for 60–90 s up to 35 cycles, with the final stabilization step carried out at 72°C for 10 min. The amplified PCR product was purified with a QIAquick PCR purification kit (Qiagen Inc., Valencia, CA, USA), and all procedures followed the supplier's instructions.
DNA sequencing and alignment
The PCR products were directly sequenced using an automatic sequencer, in this case an ABI Prism 3730xl DNA analyzer (Applied Biosystems, Foster City, CA, USA). Sequence alignments for each marker were performed using MUSCLE (multiple sequence comparison by log-expectation) (Edgar, 2004), which is implemented in Geneious Prime v. 2022.1.1 (Biomatters Ltd., Auckland, New Zealand) (Kearse et al., 2012). Subsequently, further alignment was conducted using MAFFT (Katoh et al., 2002), as needed, and the alignment was finally completed by manual adjustment.
Phylogenetic analyses
A total of 2,448 concatenated sequences of the eight plastid markers were used for the phylogenetic analyses (Table 1). Information about the aligned sequence lengths and the sequence divergence values of each marker are shown in Table 1. The validity of the sequence concatenation process was tested via an incongruence length difference test (Farris et al., 1994), in which the p-value (p = 0.001) for the partition homogeneity test was calculated using PAUP ver. 4.0a147 (Swofford, 2002) (performed with 1,000 replicates, simple addition sequence replicates, MulTrees, and TBR branch swapping, gaps treated as missing).
The phylogenetic tree was reconstructed using the Bayesian inference (BI) method. The BI analysis was conducted using MrBayes v3.2.2 (Ronquist and Huelsenbeck, 2003). jModelTest 2.1 (Guindon and Gascuel, 2003; Darriba et al., 2012) was used to evaluate the appropriate evolution model of the data combined with each dataset. The optimal model search for the BI analysis was selected using the Akaike information criterion (Akaike, 1974) method. Two independent analyses were performed using four Markov chain Monte Carlo = simulations, applying the general time reversible model, the gamma distribution of substitution rates (G), and estimates of invariant characters (I). Analyzing 10,000,000 generations, we sampled the trees every 200 generations and removed 25% of the sampled trees by burn-in processing. After burn-in removal, we calculated the 50% majority-rule sum tree and posterior probability (PP) using the remaining tree counts. The analyzed phylogenetic trees were confirmed by FigTree ver. 1.4 (Rambaut, 2012). Bootstrap percentage (BP) values for each clade were calculated to provide additional information on the reliability of the clades observed in the BI tree. To do this, a maximum parsimony heuristic search was repeated 1,000 times using PAUP ver. 4.0a147 (Swofford, 2002) with the simple addition sequence replicate, tree-bisection-reconnection branch swapping, MulTrees, and ACCTRAN options.
RESULTS AND DISCUSSION
Subfamilial phylogenetic relationships
The subfamilial phylogenetic relationships in the BI tree were basically identical to those in previous results (Bouchenak-Khelladi et al., 2010; Edwards and Smith, 2010; Grass Phylogeny Working Group II, 2011; Wu and Ge, 2012; Cotton et al., 2015; Soreng et al., 2015), recognizing three small basal subfamilies (Anomochlooideae, Pharoideae, Puelioideae) and two large and strongly supported groups: the BOP and PACMAC clades (Fig. 2). All members of Korean Poaceae were included in the large clades; therefore, all subsequent discussions on the phylogenetic relationships of Poaceae will focus on the two major clades.
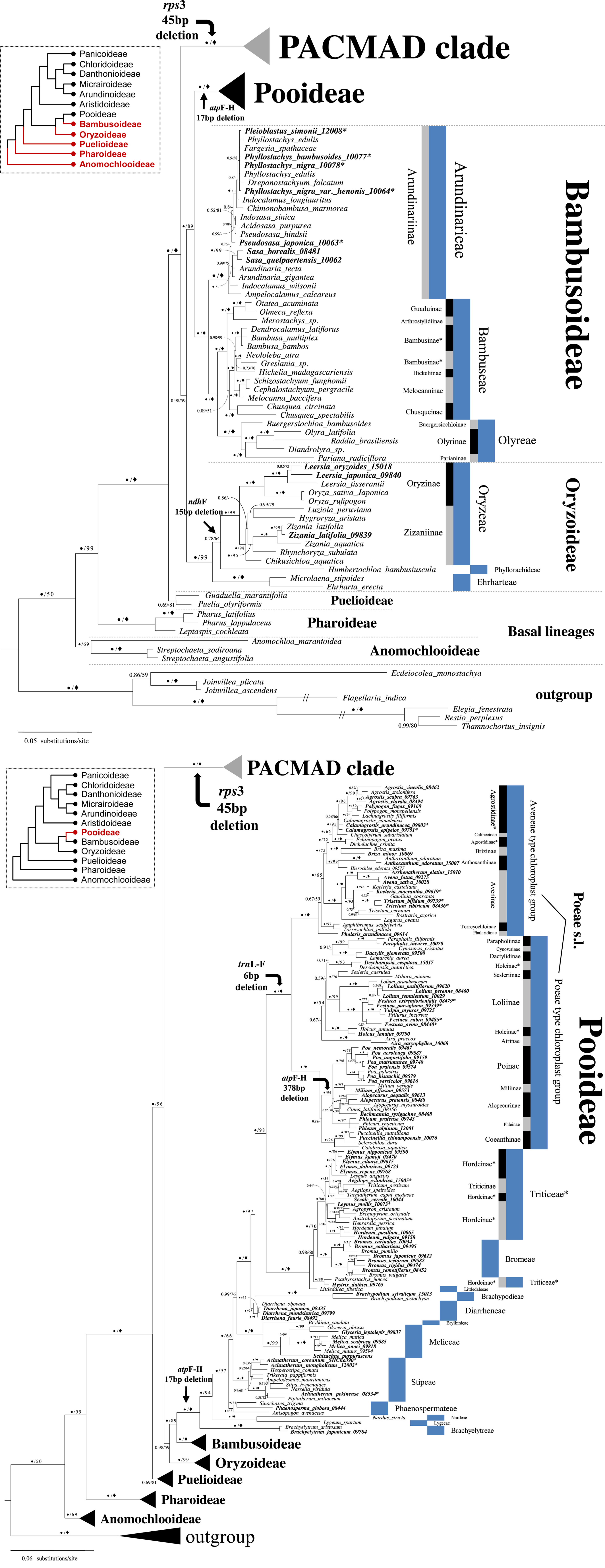
Bayesian consensus tree based on the combined sequences of eight markers, emphasizing the phylogenetic relationships of the BOP (Bambusoideae, Oryzoideae, Pooideae) clade. Posterior probabilities (● = 1.00) and bootstrap value (◆ = 100) are shown above or below. Korean taxa are in bold. Taxa with asterisks are either polyphyletic or paraphyletic. The tribal (blue) and subtribal (black or gray) classifications are from Soreng et al. (2015).
Phylogenetic relationships in the BOP clade
The phylogenetic relationships within the BOP clade differ depending on the author. A study by Gaut (2002), who estimated evolutionary patterns using the chromosome number and genome size of Poaceae, suggested a close relationship between Oryzoideae and Bambusoideae. In contrast, Mathews et al. (2000) found that Oryzoideae was more closely related to Pooideae based on the phytochrome B gene phylogeny employing 51 Poaceae taxa. In the present study, Bambusoideae and Pooideae formed a strong sister group with 1.00 PP and 89 BP support values (Fig. 2). This result is consistent with recent phylogenetic studies of BOP clades based on chloroplast genomes (Wu and Ge, 2012). Currently, the relationships among the subfamilies of the BOP clade identified in this study seem to be relatively firmly established (Hilu et al., 1999; Zhang, 2000; Salamin et al., 2002; Bouchenak-Khelladi et al., 2008, 2010; Saarela and Graham, 2010; Grass Phylogeny Working Group II, 2011).
Korean members of Poaceae belonging to the BOP clade were classified into 12 tribes (Fig. 2). The phylogenetic relationships and classification of each subfamily are as follows.
Oryzoideae: Oryzoideae was grouped into the three monophyletic tribes of Oryzeae, Phyllorachideae, and Ehrharteae. It is noteworthy that the “Oryzeae + Phyllorachideae” clade shared a 15 bp deletion in ndhF, although the clade received rather weak support (PP 0.77, BP 64). Two Korean genera, Leersia and Zinania, within tribe Oryzeae formed distinct clades in the two subtribes of Oryzinae and Zizaniinae, respectively (Fig. 2).
Bambusoideae: Bambusoideae consists of three tribes: Arundinarieae, Bambuseae, and Olyreae. All Korean taxa (representing four genera) included in this subfamily are nested in Arundinarieae. Among the four genera, Sasa forms a monophyletic group, while Phyllostachys, Pleioblastus, and Pseudosasa were found to be polyphyletic groups, exhibiting unclear interspecies relationships (Fig. 2). Chloroplast DNA phylogeny of temperate bamboo identified six major clades (the Bergbambos clade, Oldeania clade, Chimonocalamus clade, Shibataea clade, Phyllostachys clade, and Arundinaria clade) (Triple and Clark, 2010; Stapleton, 2013) within the subfamily, but the intergeneric and interspecies relationships within each clade remain not well clarified. In the present phylogeny, Korean taxa are divided into the Phyllostachys clade and Arundinaria clade, but monophyletic or phylogenetic relationships among the genera or species within each clade are not evident. This is thought to be due to the active interspecific gene flow by hybridization that is known to occur in a significant number of temperate bamboos (Triple and Clark, 2010). Low sequence variations of the markers used may also be another reason for the weak relationship, causing some incongruence between the previous morphological classification and the molecular phylogeny. It will be necessary to obtain a large amount of gene sequence data to clarify the taxonomic positions of the Korean taxa. A phylogeny based on genome-wide SNP analyses would be helpful to resolve this problem.
Pooideae: This marks the largest and most strongly supported subfamily in the BOP clade. A 17 bp deletion in atpF-H was newly observed in this study, providing additional evidence of the monophyly of the subfamily (Fig. 2). The Korean members (39 genera) of Pooideae included in this study were nested in 18 subtribes representing ten tribes. It should be emphasized that there were two additional deletions, which could indicate the robustness of certain tribal or subtribal relationships (Fig. 2). These were a trnL-F 6 bp deletion and atpF-H 378 bp deletion, supporting a monophyletic Poeae s.l. and a clade that consisted of four subtribes (Phleinae, Alopecurinae, Miliinae, and Poinae), respectively. Most of the Korean taxa were monophyletic except seven genera (Achnatherum, Leymus, Festuca, Trisetum, Koeleria, and Calamagrostis), which were found to be either paraphyletic or polyphyletic.
Achnatherum (exclusively represented by three Korean species in this study) was polyphyletic within tribe Stipeae as A. coreanum (≡Patis coreana (Honda) Ohwi) clustered with A. mongholicum (= Ptilagrostis mongholica (Turcz. ex Trin.) Griseb.), while A. pekinense (≡Stipa pekinensis Hance) formed a sister group with Piptatherum miliaceum (Fig. 2). It should be noted that the taxonomic boundary of Achnatherum has been controversial, as each of the three species has been treated as a member of the three different genera of Stipa, Patis, and Ptilagrostis, respectively (Clayton and Renvoiz, 1986; Tzvelev, 1989; Barkworth, 2007; Romaschenko et al., 2010, 2011). Expanded taxon sampling and data will clarify the taxonomic identity of Achnatherum.
Leymus in tribe Triticeae was found to be polyphyletic. Leymus mollis, a Korean accession, formed a clade with the group containing Hordeum, among others, whereas L. angustus appears as a sister species of the Elymus clade (Fig. 2). Leymus is known as one of the genera that underwent severe polyploidization in tribe Triticeae (Sha et al., 2010). Phylogenetic analyses and morphological investigations with expanded taxon sampling including L. chinensis and L. secalinus, which are known to be distributed on the Korean Peninsula, are needed to confirm their phylogenetic relationship.
In the subtribe Loliinae, Festuca formed a paraphyletic group. This is consistent with the results of molecular phylogenetic studies of this festucoid group (Torrecilla and Catalán, 2002; Torrecilla et al., 2004). Some species of Festuca have shown a close relationship with taxonomically controversial genera such as Psilurus, and Vulpia (Darbyshire and Warwick, 1992; Charmet et al., 1997; Gaut et al., 2000). Some Festuca species form a clade with Psilurus and Vulpia (Fig. 2). These Festuca species have been suggested as polyploids, and the relationships among them have not been clearly resolved. Additional data from chromosomal and phylogenetic analyses using expanded samples would be necessary to clarify the taxonomic issue.
Koeleria macrantha, a Korean accession of Koeleria, exhibited a closer relationship with Gaudinia coarctata than with K. castellana, meaning that Koeleria is a paraphyletic group. Similarly, the two Korean species of Trisetum (T. bifidum, T. sibiricum) sampled for this study did not form a clade with T. cernuum, rather the latter species showed a sister relationship with Rostraria azorica. This finding is concordant with a previous hypothesis based on nuclear internal transcribed spacer (ITS) and chloroplast DNA (trnT-H) phylogeny (Quintanar et al., 2007). Rostraria and Trisetum have been found to be very similar in terms of the epidermal characters of their lemma (Finot et al., 2006). The discrepancy between the results of molecular phylogenetic studies and the morphological traits necessitates a taxonomic reexamination of these taxa.
Lastly, monophyly of Calamagrostis within subtribe Agrostidinae was uncertain as the phylogenetic relationships among two Korean accessions (C. epigejos and C. arundinacea) and C. canadensis remains unresolved (Fig. 2). Saarela et al. (2010) found that some species of Calamagrostis form geographically segregated lineages according to nuclear and chloroplast DNA sequence data. The present study, however, did not confirm any Eurasian lineage in which the Korean accessions were included. More comprehensive analyses with additional data, especially pertaining to the nuclear genome, would be necessary to resolve the phylogenetic relationships among Calamagrostis species.
Phylogenetic relationships in the PACMAD clade
The PACMAD clade has been considered as a robust phylogenetic group since it was initially proposed by Duvall et al. (2007). The clade was also evident, being supported by the additional character of a 45 bp deletion in rps3, which was newly observed in the present study (Fig. 3). We consider the deletion to be an important diagnostic molecular trait to distinguish the PACMAD clade from all other clades of Poaceae. The subfamilial relationship was identical to the ndhF + rbcL phylogeny, which employed 448 grass species (Soreng et al., 2015). The basal position of Aristidoideae within the PACMAD clade, however, was poorly supported in the present study (0.72 PP, 70 BP). In some studies, Aristidoideae has been found to have close relationships with Danthonioideae and Chloridoideae, but such relationships have not been well supported (Hsiao et al., 1999; Grass Phylogeny Working Group, 2001; Bouchenak-Khelladi et al., 2008, 2010). Cotton et al. (2015) performed a chloroplast genome analysis of the PACMAD clade using a limited number of representative taxa and argued that Panicoideae was the first to diverge in the clade. Lee (2016) also reconstructed the plastid genome (76 coding genes) phylogeny of Poaceae with expanded taxon sampling, but the basal position Aristidoideae varies depending on the phylogenetic method used.

Bayesian consensus tree of eight combined markers, emphasizing the phylogenetic relationships of the PACMAD (Panicoideae, Arundinoideae, Chloridoideae, Micrairoideae, Aristidoideae, Danthonioideae) clade. Posterior probabilities (● = 1.00) and bootstrap value (◆ = 100) are shown above or below. Korean taxa are in bold. Taxa with asterisks are either polyphyletic or paraphyletic. The tribe (blue) and subtribes (black or gray) are from Soreng et al. (2015).
Our study supported the subfamilial delimitation and relationships observed in previous studies (Clark et al., 1995; Mathews and Sharrock, 1996; Hilu et al., 1999; Sánchez-Ken et al., 2007; Vicentini et al., 2008; Grass Phylogeny Working Group II, 2011; Peterson et al., 2011; Soreng et al., 2017; Saarela et al., 2018). Among the six subfamilies in the PACMAD clade, Korean Poaceous taxa were classified into five subfamilies (excluding Aristidoideae) and 11 tribes (Fig. 3). The phylogenetic relationships and taxonomic considerations within each subfamily are described below.
Arundinoideae: All Korean accessions representing Phragmites (P. australis, P. japonicus) and Molinia (M. japonica) were nested in a highly robust subtribe, Molinieae (Fig. 3). The members of Molinieae shared a noteworthy 98 bp indel in atpF-H, possibly serving as a useful molecular marker defining the subtribe. The close relationship between the two genera is confirmed in the nuclear ribosomal internal transcribed spacer (nrITS) phylogeny based on a single accession for each genus (Hsiao et al., 1998). Two Korean accessions of Phragmites formed a clade with P. mauritianus. However, M. japonica formed a monophyletic group with Phragmites spp., and not with M. caerulea, making Molinia possibly paraphyletic. Additional taxon sampling and data will be necessary to clarify the taxonomic circumscription and delimitation of the two genera further.
Micrairoideae: This subfamily includes three Korean species representing Coelachne and Isachne included in the subtribe Isachneae. Of the genera, monophyly of Isachne was evident as the two Korean accessions were closely allied with I. distichophylla (Fig. 3). It should be emphasized that these two genera have previously been included in Panicoideae (Clayton and Renvoize, 1986; Watson et al., 1992) and Chloridoideae as well (for example C. japonica) (Lee, 2007). The present study suggests that the Korean species should be included in Micrairoideae.
Danthonioideae: There is no Korean native species included in this subfamily. The newly examined accession representing the subfamily is Cortaderia selloana, which has been introduced from South America for landscaping. There is another introduced species Danthonia spicata, which distributes in the middle part of Korea (Lee, 2016), but is not included in this study. Reimer (2006) studied the interspecific relationship of Danthonia using the trnL-F, confirming that D. spicata showed the closest relationship to D. parryi and D. unispicata.
Chloridoideae: The Korean accessions representing seven genera were classified into three tribes (Cynodonteae, Eragrostideae, Zoysieae) and five subtribes (Eleusininae, Eragrostidinae, Muhlenbergiinae, Sporobolinae, Zoysiinae) in Chloridoideae (Fig. 3). Among the seven genera, Eragrostis did not form a monophyletic group. Some of the species of Eragrostis showed a close phylogenetic relationship with Pogonarthria and Ectrosia, making Eragrosits polyphyletic. Similar results have been obtained from phylogenetic studies using the sequences of nrITS and trnL-F (Columbus et al., 2007) and waxy and rps16 (Ingram and Doyle, 2004). Additional studies including samples of E. multicaulis, E. ferruginea, and E. japonica will clarify the taxonomic circumscription of the genera.
Panicoideae: The taxa previously classified as ‘Centothecoideae’ were included in Panicoideae (Fig. 3). These taxa had been classified as Arundinoideae based on morphological and anatomical investigations (Renvoize, 1981; Hilu and Wright, 1982; Davis and Soreng, 1993; Hsiao et al., 1998). Grass Phylogeny Working Group (2001) and Bouchenak-Khelladi et al. (2008) argued that ‘Centothecoideae’ was a monophyletic group which could be classified as a distinct subfamily, but several molecular phylogenetic studies have shown that the members of ‘Centothecoideae’ have closer relationships with Panicoideae (Hsiao et al., 1999; Zhang, 2000; Grass Phylogeny Working Group, 2001; Salamin et al., 2002; Duvall et al., 2007; Sánchez-Ken and Clark, 2007; Bouchenak-Khelladi et al., 2008, 2010). Sánchez-Ken and Clark (2007, 2010) and Grass Phylogeny Working Group II (2011) have also argued that the separation of ‘Centothecoideae’ as an independent subfamily was not appropriate, a contention supported by the present study. It is thought that the monophyly of 'Centothecoideae' observed in previous studies (Grass Phylogeny Working Group, 2001; Bouchenak-Khelladi et al., 2008) is due to the limited taxon sampling in those studies.
The Korean accessions (representatives of a total of 28 genera) included in this subfamily were nested in five tribes (Zeugiteae, Paniceae, Paspaleae, Arundinelleae, and Andropogoneae) (Fig. 3). Except for the accessions of three monophyletic genera (Lophatherum, Paspalum, and Arundinella) belonging to the relatively small tribes Zeugiteae, Paspaleae, and Arundinelleae, the remaining Korean accessions were divided into two large tribes, Paniceae (9 genera) and Andropogoneae (16 genera). Monophyly of many genera within these two large tribes is not evident, as discussed below.
Within the subtribe Cenchrinae (tribe Paniceae), Pennisetum and Setaria were found to be polyphyletic, as some members of each genus were grouped with species of other genera. Chemisquy et al. (2010) suggested merging Pennisetum into Cenchrus based on the results of morphological and molecular phylogenetic analyses. Meanwhile, P. glaucum, which showed a sister relationship with S. pumilia, was sometimes recognized as S. glauca (Hus, 1978; Lee, 2007). It is apparent from the molecular phylogeny that P. glaucum is distinct from other species in the genus, but the taxonomic treatment must be verified as the generic circumscriptions of related taxa have not yet been fully resolved. For example, Setaria also appears to be polyphyletic (Fig. 3). Polyphyly of the genus has also been suggested in molecular phylogenetic analyses and inflorescence differentiation pattern studies (Doust and Kellogg, 2002; Doust et al., 2007; Kellogg et al., 2009). Korean Setaria species are divided into two groups; the first is an annual, with highly condensed panicle and spikelets with bristles (S. chondrachne, S. pycnocoma, S. verticillata, S. italica, S. viridis); the second group is an annual or perennial, with loose panicles, narrow spikelets, and wrinkled leaves (S. pumila). This is consistent with the results reported by Kellogg et al. (2009).
Eriochloa in the subtribe Melinidinae is paraphyletic, and the two representative species of the genus form a clade with Urochloa panicoides (Fig. 3). These two genera can be distinguished by the shape of the callus, i.e., the base of the spikelets; Eriochloa has a swollen cup shape, whereas Urochloa does not (Salariato et al., 2009). However, the other morphological features are very similar. Molecular phylogenetic studies using nuclear ITS sequences also did not support monophyly in Eriochloa (Torres Gonzalez and Morton, 2005). Because only one species (E. villosa) is currently distributed in Korea, a phylogenetic analysis including expanded taxon sampling from the genus as well as closely related genera would be necessary to address the taxonomic issue.
Two Korean accessions of Digitaria in the subtribe Anthephorinae are divided into two different clades: D. sanguinalis shows a sister group relationship with the “Anthephora elongata + Chaetopoa pilosa” clade and D. violascens is allied with Megaloprotachne albescens (Fig. 3). It is interesting to note that a close relationship between these genera and Digitaria has not yet been proposed. The two Korean species can be distinguished by the surfaces of the spikelets (smooth vs. the presence of protrusions) (Vega et al., 2009). Given that the phylogenetic positions of the two species are very different from each other, a more comprehensive study should be conducted to clarify the generic identity of Digitaria.
The Korean accessions classified as the subtribe Panicinae were Panicum bisulcatum, P. acuminatum, and P. dichotomiflorum, which were found to be polyphyletic (Fig. 3). To date, many taxa within Panicum have been transferred to other genera or reclassified into new genera based on morphological traits and molecular phylogenetic studies (Aliscioni et al., 2003; Bess et al., 2006; Morrone et al., 2007, 2008, 2012; Sede et al., 2008, 2009; Scataglini and Zuloaga, 2013). The three Korean taxa included in Panicum were placed at different positions in the phylogenetic tree. However, evident morphological characters to distinguish them have not yet been found. Washburn et al. (2015) reported that P. bisulcatum and P. acuminatum are C3 plants, while P. dichotomiflorum will perform C4 photosynthesis according to a comparative analysis of photosynthesis types. It appears that more detailed morphological, physiological, and molecular phylogenetic investigations including Panicum and all relevant genera are required.
The Korean taxa classified as tribe Andropogoneae were divided into 16 genera, and their subtribal classification and interspecies relationship were found to be very complicated. Only two subtribes (Arthraxoninae and Coicinae) were monophyletic, while all other subtribes (Ischaeminae, Dimeriinae, Rottboelliinae, Saccharinae, Anthistiriinae, and Sorghinae) were found to be either paraphyletic or polyphyletic (Fig. 3). Most of the Korean taxa belonging to this tribe are also polyphyletic. This result is consistent with the findings of a phylogenetic study using ITS and trnL-F (Skendzic et al., 2007). Andropogoneae is known to diversify in a very short period, causing difficulty in establishing a stable classification system. Several studies have attempted to elucidate the phylogeny of this tribe using various data, but no meaningful progress has been made thus far. This is presumably because these taxa have undergone severe evolutionary radiation and frequent hybridization (Kellogg, 2000; Mathews et al., 2002; Skendzic et al., 2007; Doyle et al., 2008; Flagel and Wendel, 2010; Teerawatananon et al., 2011). To elucidate their relationship in more detail, it is thought that a large amount of genome-wide SNP data as well as intensive research on the major morphological traits that can be used to classify each subtribe are needed.
Acknowledgements
This study was supported by a grant from the National Institute of Biological Resources of the Republic of Korea (NIBR202212101).
Notes
CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest.
References
Appendices
Taxa employed for the phylogenetic analyses of Poaceae.
kjpt-52-3-127-app.pdf
