Michikwangipul, Scopolia parviflora (Dunn.) Nakai (Dunn, 1912; Nakai, 1933) of the family Solanaceae, limitedly occurs in Korean peninsula, and is similar to S. japonica (Maximowicz, 1873) occurring in Japan. S. parviflora is distinguished from S. japonica by ITS and some phenetic characters (Hong and Paik, 2001; Kim et al., 2003). The members of the genus Scopolia are known as medicinal plants (Mino, 2002; Jung et al., 2003; Min et al., 2007). The genetic makeup of the plastid genome of Scopolia is poorly known. Here we sequenced the chloroplast genome of Scopolia parviflora as a representative of the genus Scopolia.
Materials and Methods
The plant material of Michikwangipul used in this study was collected from the wild population of Mt. Cheonma, Korea (N37° 40′ 34.85″ E127° 15′ 33.00″). The voucher of the plant specimen (Parkjh 20150505-141) was deposited in NNIBR, Herbarium in Nakdonggang National Institute of Biological Resources. The total DNA was prepared as described by Lee et al. (2015). The Illumina paired-end (PE) genomic library of 200 bp was constructed and sequenced using an Illumina HiSeq 2000 platform. The plastid sequence was obtained using CLC Genomics Workbench version 7.05 as described by Jeong et al. (2014). Circular structures of each replicon were confirmed by polymerase chain reaction (PCR) amplification at their ends and by joining of Sanger sequence reads derived from the amplicons. The assemblies were further verified by examining paired-end distance and depth after re-mapping reads on the contig sequences. The BLAST searches of a large contig were verified to be plastid genomes. For gene annotation of organelle genomes, protein-coding and ribosomal RNA genes were annotated using DOGMA (http://dogma.ccbb.utexas.edu/; Wyman et al., 2004). The boundaries of each annotated gene were manually determined by comparison with orthologous genes from other known cp genomes. Genes encoding tRNAs were first predicted using tRNAscan (http://lowelab.ucsc.edu/tRNA scan-SE; Lowe and Eddy, 1997) and ARAGORN version 1.2 (http://130.235.46.10/ARAGORN/; Laslett and Canback, 2004), and were manually verified by predicting the tRNA secondary structure. Circular genome maps were drawn using GenomeVx (Conant and Wolfe, 2008) followed by manual modification. The sequencing data and gene annotations were submitted to GenBank with accession number KU900232. DNA sequences of seven cp protein genes, including psaA, psaB, psbA, psbB, psbC, psbD, and rbcL were used to construct cp phylogenetic tree by Maximum Parsimony criterion using Paup ver. 6.0. Bootstrap and jackknife analyses of the MP tree were also performed with 1,000 replicates.
Results and Discussion
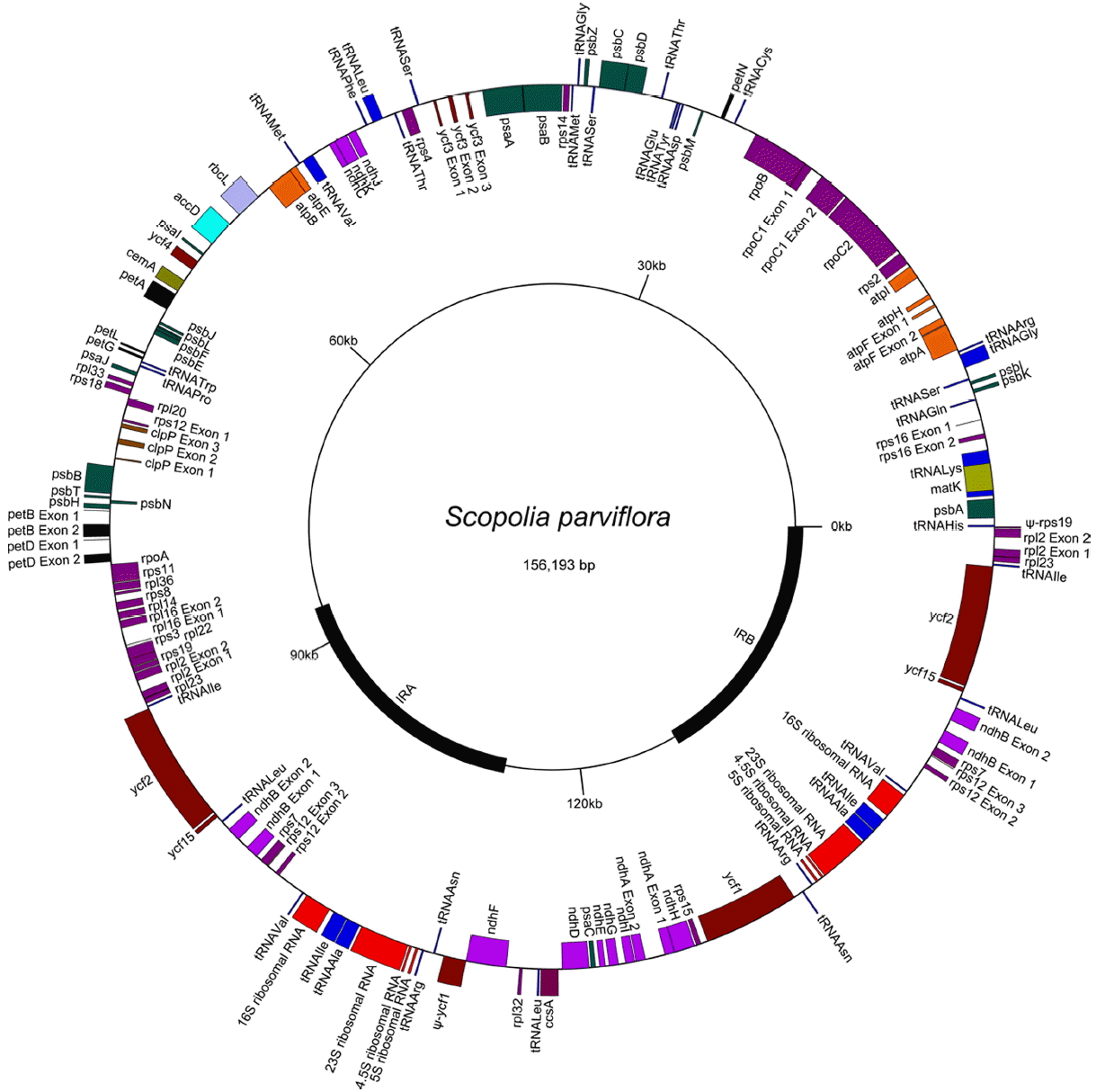
The cp-genome of Scopolia parviflora, was determined (Fig. 1) and found to be 156,193 bp in length. It includes small and large single copy (SSC, LSC) regions of 18,019 bp and 86,364 bp, respectively, separated by a pair of 25,905 bp inverted repeats (IRs). A total of 113 genes were detected, including 80 protein coding genes, 30 tRNA genes, and four rRNA genes (Table 1). This cp-genome was also found to contain 21 different introns, including 20 group II introns and a group I intron with a cyanobacterial origin (Besendahl et al., 2000) found within the trnL_uaa gene. Three protein coding genes, including clpP, rps12, and ycf3, contain two group II introns (clpP.i1, clpP.i2, rps12.i1, rps12.i2, ycf3.i1 and ycf3.i2), and 14 genes contain a single group II intron: rpoC1.i, rpl2.i, rpl16.i, rps16.i, atpF.i, petB.i, petD.i, ndhA.i, ndhB.i, trnA_ugc.i, trnG_ucc.i, trnI_gau.i, trnK_uuu.i, and trnV_uac.i. Among the 20 group II introns, the intron in rps12, between exons 1 and 2, is trans-splicing, while the other 19 group II introns are cis-splicing.
Seventeen genes, five introns, and parts of three genes and an intron are found within the IR, which has two copies. These 17 genes include six protein-coding genes (ndhB, rpl2, rpl23, rps7, ycf2, ycf15), all four rRNA genes (16S, 23S, 4.5S, 5S), and seven tRNA genes (trnA_ugc, trnI_cau, trnI_gau, trnL_caa, trnN_guu, trnR_acg, trnV_gac). The five introns are ndhB.i, rpl2.i, trnA_ugc.i, trnI_gau.i, and rps12.i2. The IR contains the 5’ end of ycf1 at the border with the SSC, resulting in one intact ycf1 and a 1,473-bp ψ-ycf1 in the cp-genome. The IR also contains the 5’ end of rps19 at the border with the LSC, resulting in one intact rps19 and a 84-bp ψ-rps19 in the cp-genome. In addition, the IR contains parts of the rps12 gene. This rps12 gene consists of three exons, rps12.e1, rps12.e2, and rps12.e3, rps12.e1 is in the LSC, but rps12.e2 and rps12.e3 are in the IR. Thus, the genome contains a single copy of rps12.e1 but has two copies of rps12.e2 and rps12.e3. A cis-splicing group II intron, rps12.i2, intervenes between rps12.e2 and rps12.e3, but a trans-splicing intron, rps12.i1t, occurs between rps12.e1 and rps12.e2. The rps12.i1t is split into two pieces, rps12.i1t1 and rps12.i1t2, because the rps12 gene is transcribed in two separate operons, the clpP operon (clpP- rps12.e1- rps12.i1t1-rpl20) and the 3’ rps12 operon (rps12.i1t2-rps12.e2-rps12.i2-rps12.e3-rps7-ndhB).
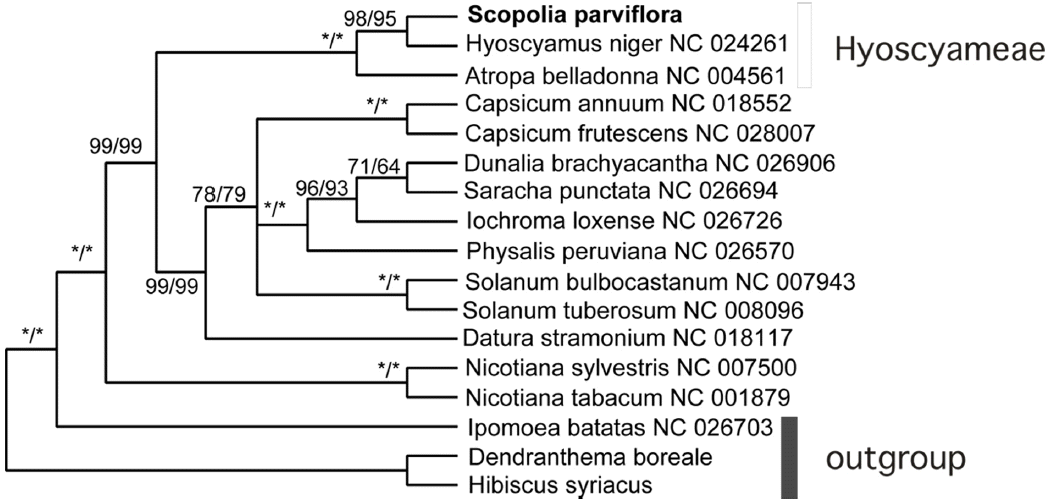
Currently, more than 20 plastid genomes have been deposited in Genbank from 10 genera of Solanaceae. Phylogenetic analysis showed that Scopolia parviflora formed a strong clade with Hyoscyamus niger (Sanchez-Puerta & Abbona, 2014) and Atropha belladonna (Schmitz-Linneweber et al., 2002), and that Hyoscyamus niger was the closest to Scopolia parviflora. The results support monophyly of the tribe Hyoscyameae of Solanaceae. The three plants are known to be highly toxic and are also used as medicine. In contrast, another toxic plant, Datura (Yang et al., 2014), claded with potato (Fig. 2).
In Korea, two species of the genus Scopolia have been documented. One is purple flowered Scopolia parviflora (Dunn.) Nakai (1933) and the other is yellow flowered S. lutescens Y. Lee (1993). In contrast, purple flowered Scopolia japonica Maxim. (1873) and yellow flowered Scopolia japonica Maxim. f. lutescens Sugim. (1977) also occur in Japan. Currently, ITS sequence analysis suggested that Scopolia parviflora and S. japonica were clearly distinguished, but that Scopolia parviflora and S. lutescens Y. Lee were indistinguishable (Kim et al., 2003). Plastid DNA sequence of Scopolia japonica from Japanese collection (voucher Tsugaru & Sawada, 17731) was only available in ndhF (Genbank EU580945). Without 162 ambiguous sequence of EU580945, Scopolia japonica had 5 SNPs in ndhF distinguished from Scopolia parviflora. Further study of comparative plastid genomics would help our understanding on the relationship among the Scopolia species.