INTRODUCTION
Viola L. is mainly distributed in temperate and tropical regions in the Northern Hemisphere and is known to contain about 658 taxa (Marcussen et al., 2022). However, the allied species are similar in morphology, and interspecific hybridization occurs frequently in the natural habitats, making distinguishing species difficult (Russel, 1960). Various studies were conducted on Korean Viola, including morphological studies (Lee and Lee, 1968; Kim, 1986; Whang, 2002), palynological and seed micromorphological studies (Lee, 2022), cytological studies (Lee, 1967, 1969), molecular phylogenetic studies based on DNA sequences (Yoo et al., 2005, 2007; Yoo and Kim, 2006; Yoo and Jang, 2010), and complete chloroplast genome sequencing for some taxa (Cheon et al., 2017, 2019, 2020; Kwak, 2021; Go et al., 2022; Moon and Kim, 2022; Park et al., 2023). However, there are few comparative studies of morphologically similar taxa (Valentine, 1950; Mohammadi Shahrestani et al., 2014). V. selkirkii Purch ex Goldie, acauline taxa, is distributed in the Northern Hemisphere and throughout Korea (GBIF Secretariat, 2022). Nakai (1909) reported its distribution in Korea for the first time, and Nakai (1919) reported its distribution in Ulleung-do Island (UE) for the first time. V. ulleungdoensis M. Kim & J. Lee is a Korean endemic species designated as a novel species showing distinctions that it is distributed only in UE, has no adventitious bud unlike V. selkirkii, and grows at low altitude with large leaves and increasing plant size after flowering (Lee et al., 2012). However, studies on V. selkirkii and V. ulleungdoensis have revealed controversial findings because the species are morphologically similar (Yang et al., 2015; Gil and Kim, 2016). A study on the flora in UE (Yang et al., 2015) reported the distribution of only V. selkirkii, except for V. ulleungdoensis; therefore, the two species seem to be considered the same. In a study about V. woosanensis (Gil and Kim, 2016), V. ulleungdoensis was contributed as the maternal parent for several reasons and referred to as “previously recognized as V. selkirkii,” thereby categorizing V. selkirkii and V. ulleungdoensis from UE as the same species. In terms of external morphology, there has been a controversy about identifying the two species of V. selkirkii and V. ulleungdoensis growing in UE because they have similar characteristics, including glabrous petioles and peduncle, beardless petals, purple flowers, and capitate stigma (Lee et al., 2012). Therefore, we compared two morphologically similar species through chloroplast genome sequencing, which is an easy method to compare similar taxa because of easy DNA extraction and high homology of gene structure and sequence (Simpson, 2019).
MATERIALS AND METHODS
V. selkirkii, which is distributed throughout South Korea, was collected for comparison of intraspecific variation from Jeju-do Island (JJ), Hwacheon-gun in Gangwon Province (HC), and V. ulleungdoensis collected. Based on the taxonomic characteristics and geographic distribution of the two species Lee et al. (2012) identified as novel species, one individual of each was collected (Table 1). The materials used in the experiment were made specimens with voucher numbers and deposited in Kangwon National University Herbarium (KWNU).
DNA extraction was performed using DNeasy Plant Mini Kit (Qiagen, Hilden, Germany), and for the sequences 3,364,589–8,012,029, raw reads were obtained at 301 bp paired-ends using the Illumina MiSeq platform (San Diego, CA, USA). For de-novo assembly, we first collected chloroplast-related 208,191–496,496 reads with reference to V. seoulensis Nakai (GenBank No. KP749924) using the Map to Reference tool of Geneious 7.1.9 (Biomatters Ltd., Auckland, New Zealand). The chloroplast genome sequences were annotated using GeSeq (Tilich et al., 2017), and the annotations were manually reviewed and modified compared with the previously reported chloroplast genome sequences of V. seoulensis (KP749924), V. mirabilis (MH229816), V. websteri (MH229819), and V. japonica (MT012304) in Viola. tRNA was further identified using tRNAscan-SE 2.0 (Chan and Lowe, 2019). For sequence comparison, we used mVISTA (Frazer et al., 2004), which can identify the sequence by aligning the long-read DNA genomic sequences and visualizing these alignments. Three individuals of V. selkirkii were analyzed using V. selkirkii from UE as the reference sequence. In the comparison of four individuals (three V. selkirkii and one V. ulleungdoensis), V. ulleungdoensis was used as the reference sequence. Some regions with low sequence homology (<75%) based on the respective mVISTA results, were rearranged using MAFFT v. 7.017 (Katoh et al., 2002), and the aligned sequences were compared.
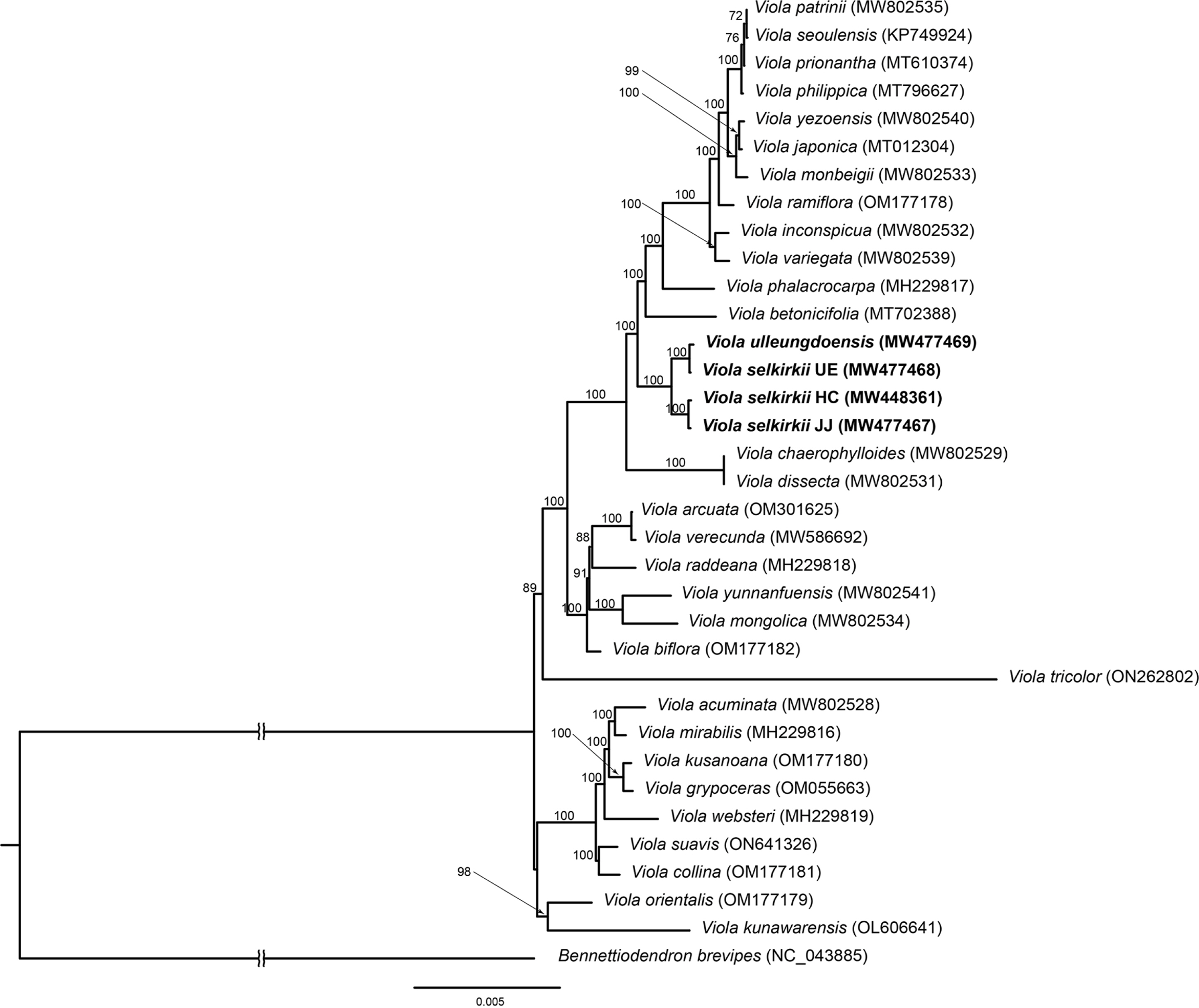
Phylogenetic analysis included 34 individuals of 32 taxa belonging to Viola, and Bennettiodendron brevipes was selected as an outgroup in Salicaceae, the family closest to Violaceae in Malpighiales (Duan et al., 2020). In phylogenetic analysis, 77 protein-coding genes common to 34 individuals were aligned using MAFFT v. 7.017 (Katoh et al., 2002) and checked manually. The aligned length was 80,864 bp. Maximum likelihood analysis was performed using RAxML-HPC2 on XSEDE v.8.2.12 (Stamatakis, 2014) with 1,000 bootstrap replicates using the CIPRES Science Gateway (http://www.phylo.org) (Miller et al., 2010) under the GTR+GAMMA substitution model.
RESULTS AND DISCUSSION
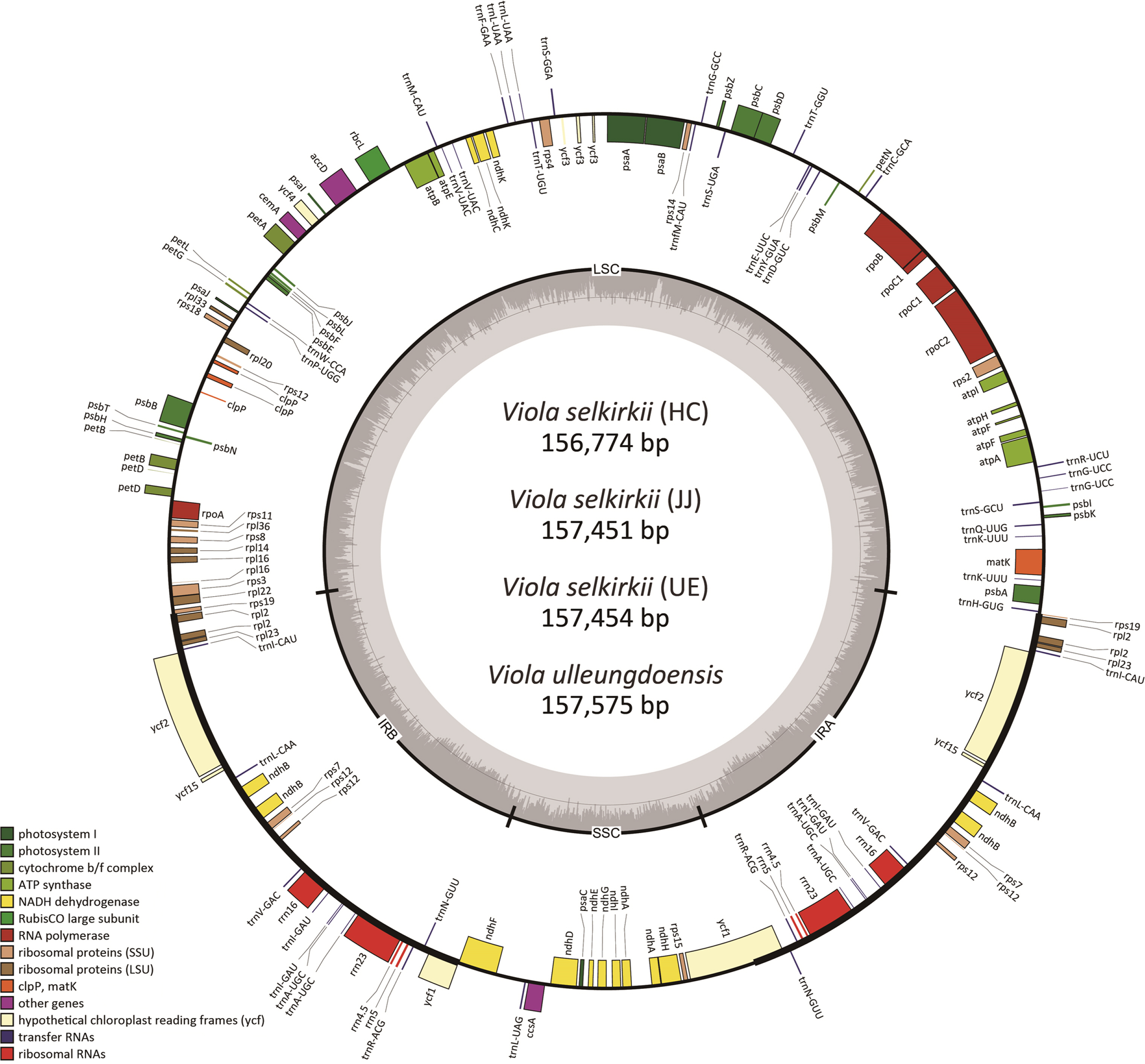
The chloroplast genome sequences of V. selkirkii and V. ulleungdoensis were found to contain 156,774–157,454 bp and 157,575 bp, respectively (Table 2). The chloroplast genome sequences of the two taxa exhibited the quadripartite structure, identical to those of the previously reported Viola, and consisting of large single-copy (LSC), small single-copy (SSC), and two inverted repeats (IR) regions (A and B) that have mirror images of each other between the two regions (Fig. 1). The LSC region of V. selkirkii was 85,921–85,950 bp in length, while that of V. ulleungdoensis was longer (86,075 bp). The SSC region of V. selkirkii was 17,261–17,982 bp in length. Among the four individuals, V. selkirkii obtained from HC had the longest SSC region (17,982 bp) and V. ulleungdoensis had the shortest SSC region (17,258 bp). The IR regions of the three individuals of V. selkirkii were 26,431–27,123 bp in length; V. selkirkii obtained from HC had the shortest IR region, while V. ulleungdoensis had an IR region spanning 27,121 bp, which was similar to the length of the IR regions of V. selkirkii obtained from JJ and UE. The GC content of the chloroplast genome was confirmed to be 36.3% in all four individuals (Table 2).
The total number of genes was 111, consisting of 77 protein-coding genes, 30 tRNAs, and four rRNAs (Table 2). In total, seven protein-coding genes (rps2, rpl23, ycf2, ycf15, ndhB, rps7, and rps12), seven tRNA genes (trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG, and trnN-GUU), and four rRNA genes (rrn16, rrn23, rrn4.5, and rrn5) were located in the IR region, and two pseudogenes (ψrps19 and ψycf1) caused by genes present across each boundary of LSC/IRa and SSC/IRb were identified. Of the 111 genes, 17 genes had introns, of which 14 genes (atpF, ndhA, ndhB, petB, petD, rpl16, rpl2, rpoC1, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) contained one intron and the remaining three genes (clpP, rps12, and ycf3) contained two introns. This structure was identified in four individuals and found to match all previously reported chloroplast genome features of Viola species, supporting the results of prior studies (Cheon et al., 2019; Chao et al., 2022) reporting that chloroplast genomes of Viola species were well preserved.
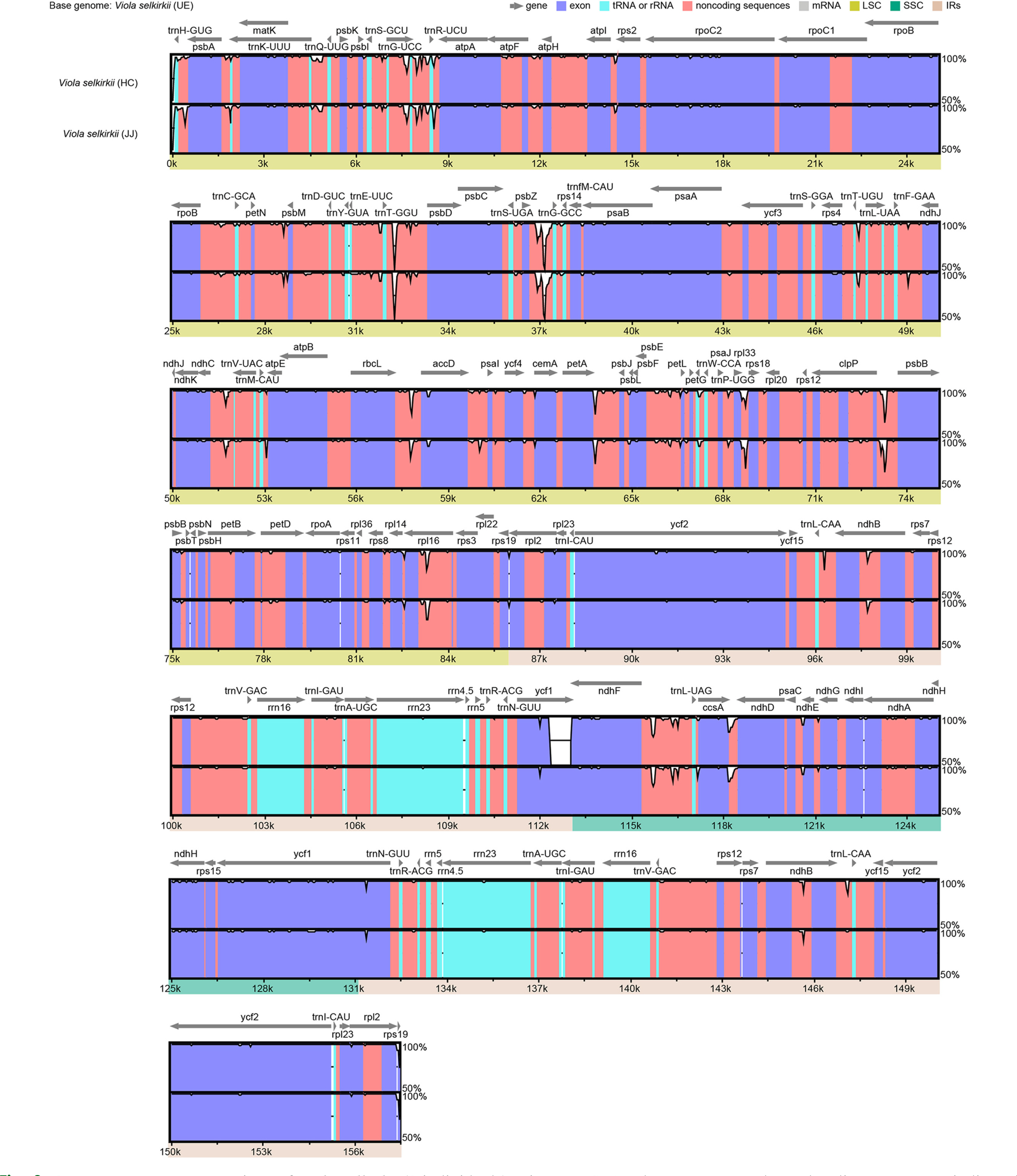
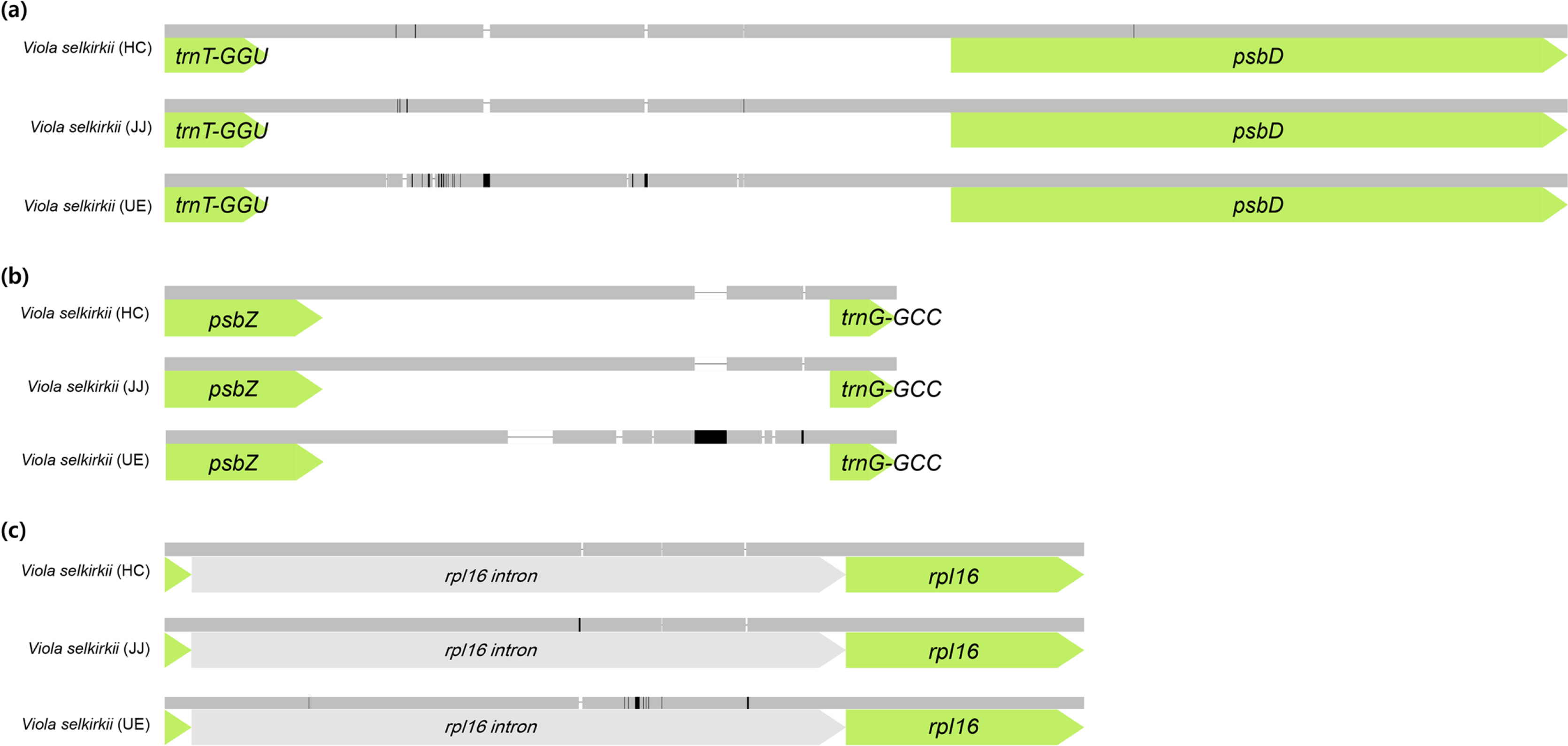
According to the results of comparing the chloroplast genome homology of three individuals of V. selkirkii using the mVISTA tool (Fig. 2), the most variable region was the intergenic spacer region. In particular, four indels and two single-nucleotide polymorphisms (SNPs) were identified in the trnT-GGU-psbD region, and seven indels were included in the psbZ-trnG-GCC region. Among the coding genes, the rpl16 gene had three indels and 16 SNPs, showing the lowest homology (Fig. 3). Except for one indel (1 bp) identified in the trnT-GGU-psbD region of 32 variable regions (14 indels and 18 SNPs), remaining 31 variable regions were the same in the individuals from HC and JJ, while they were clearly distinguished from those of the individuals from UE.
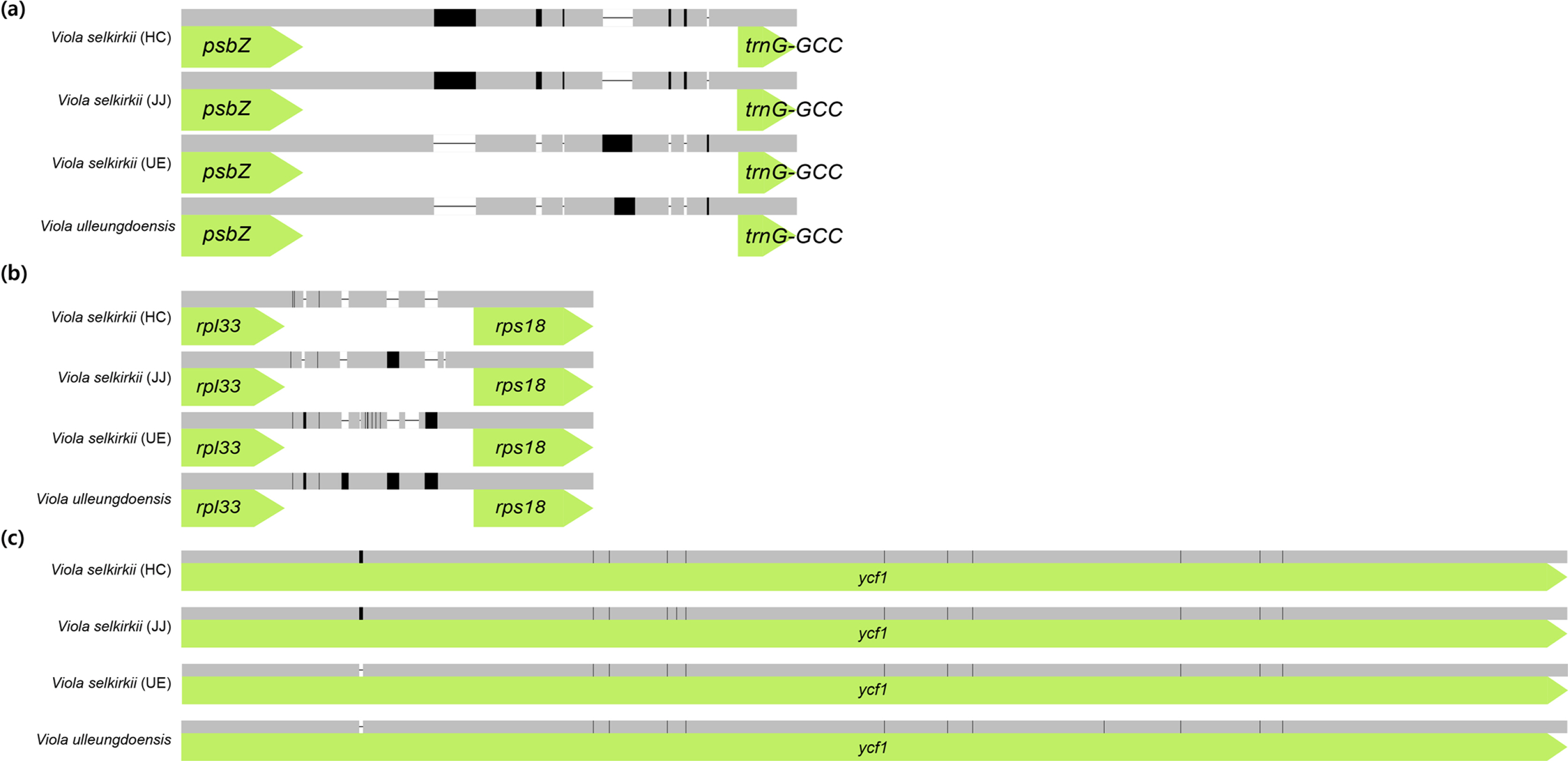
Comparison of the homology of interspecific chloroplast genome sequences of V. selkirkii and V. ulleungdoensis (Fig. 4) revealed psbZ-trnG-GCC and rpl33-rps18 as the regions with the lowest similarity. Based on this result, the sequence was realigned using MAFFT v. 7.017 (Katoh et al., 2002). Consequently, seven indels were identified in the psbZ-trnG-GCC region and five indels and nine SNPs were identified in the rpl33-rps18 region. Among the coding genes, one indel and 12 SNPs were identified in ycf1, indicating the lowest sequence homology (Fig. 5). Of the 34 variable regions (13 indels and 21 SNPs) identified above, only one SNP was found in the V. selkirkii individual from HC and one indel and one SNP were found in that from JJ. On the other hand, V. selkirkii from UE had seven variable regions including one indel and six SNPs. In the comparison based on native habitat, individuals of V. selkirkii growing in HC and JJ were differentiated in 22 variable regions (10 indels and 12 SNPs), compared to V. selkirkii and V. ulleungdoensis growing in UE. Moreover, the only variable regions that had the same sequence among three individuals of V. selkirkii and were distinguished from V. ulleungdoensis were one indel of rpl33-rps18 and one SNP of ycf1. In particular, ycf1 is valued as a DNA barcode for the classification of land plants due to its high sequence variability (Dong et al., 2015).
Meanwhile, according to the mVISTA results, a region with 0% homology by pseudo ycf1 (ψycf1) at the boundary of SSC/IRb was identified in V. selkirkii from HC (Figs. 2, 3). This region was about 700 bp shorter than the regions in other individuals, which resulted from the contracted IR region and the expanded SSC region compared to other individuals. Although contraction and expansion of these IR/SSC regions have been reported at the species or genus level (Plunkett and Downie, 2000; Zhu et al., 2016; Ni et al., 2017; He et al., 2020; Guo et al., 2021), and identified in individual cultivar species (Goulding et al., 1996). When we compared the ψycf1 length of Viola in the NCBI database, V. japonica and V. philippica also differed among individuals.
Furthermore, phylogenetic analysis revealed that 34 individuals of Viola formed a monophyletic group (BS = 100), and the four individuals investigated in this study also formed a monophyletic group (BS = 100) (Fig. 6). Among them, two individuals of V. selkirkii from HC and JJ and V. selkirkii from UE and V. ulleungdoensis each formed a subclade with high support (BS = 100).
In summary, Lee et al. (2012) used altitude and plant size as characteristics to distinguish V. ulleungdoensis and V. selkirkii in UE. However, when we conducted field research, the two species according to altitude were not clearly distinguished, and the plant sizes were classified as large or small but appeared continuously. In addition, we surveyed the field that V. ulleungdoensis is larger in plant size than the continental species of V. selkirkii, it might be an example of insular gigantism in which the size of leaves or flowers increases in UE (Yang et al., 2019a, 2019b; Park and Park, 2020). As a result of this study, V. selkirkii and V. ulleungdoensis in UE are most similar than the two species of V. selkirkii growing in HC and JJ. These results support the study of Gil and Kim (2016) in that V. selkirkii in UE and V. ulleungdoensis are recognized as the same species.