Abstract
Halenia coreana is an endangered, endemic species that is distributed in only a few locations in Korea, such as Mts. Hwaaksan and Daeamsan. It has been recently segregated from H. corniculata, broadly distributed in cold temperate regions that include northern Japan, the Russian Far East, northeastern China, Mongolia, and eastern Europe, where population sizes are usually large. To examine the genetic diversity of H. coreana and evaluate the level of genetic differentiation of the species compared with that of H. corniculata, we surveyed 183 candidate simple sequence repeats (SSR) motif markers for H. coreana and H. corniculata from sequence data of amplified fragments of a specific length in the genome. A total of 17 genomic-SSR markers were selected to examine the levels of genetic diversity and differentiation using 17 samples of H. coreana and 60 samples of three populations of H. corniculata. The results here suggest that the genetic diversity of H. coreana is very low with a high frequency of inbreeding within its population. We found that H. coreana is genetically differentiated from H. corniculata, supporting the recognition of the geographically isolated H. coreana as a distinct species.
INTRODUCTIONThe discovery of new plant species has increased over the past two decades amid a global crisis in biodiversity (Donoghue and Alverson, 2000; Kim and Byrne, 2006; Cheek et al., 2020). Many of these new species show a limited distributional range. They were found from extensive fieldwork in remote areas that were previously inaccessible or rarely visited (Kim and Byrne, 2006; Sobral and Stehmann, 2009; Do et al., 2019; Kim et al., 2019; Kang et al., 2021) or from detailed molecular and morphological analyses, in which a new species was a collection of a few populations that had been recognized as part of more widely distributed species (Chung et al., 2017; Choi et al., 2019; Han et al., 2019; Nam et al., 2020). These newly described species are often endemic to a certain area with a small number of populations that are not thoroughly evaluated on conservation lists such as International Union for Conservation of Nature’s red list or a similar list at the local level (National Institute of Biological Resources, 2012). The risk of the extinction of endemic species given their small population sizes is much higher than that of widespread species due to genetic drift, inbreeding depression, and anthropogenic activities. In addition, the probability of the extinction of endemic species induced by climate change is much higher than this probability for non-endemic species (Urban, 2015) such that more attention should be directed toward narrow endemic species. Studies of genetic diversity and population structures of endemic species with narrow distributions are particularly important to implement specific conservation strategies.
Halenia coreana S. M. Han, H. Won & C. E. Lim (Gentianaceae) is a recently described species endemic to Korea (Han et al., 2019). This biennial herbaceous species is only known from a few localities with a small number of individuals and is designated as an endangered species in Korea (https://species.nibr.go.kr/endangeredspecies/). Before a detailed morphological and molecular study of the species (Han et al., 2019), plants of H. coreana were recognized as H. corniculata (L.) Cornaz widely distributed in northeastern China, Mongolia, northern Japan, Sakhalin, the Kuriles, Kamchatka, and eastern Europe (Toyokuni and Yamazaki, 1993; Ho and Pringle, 1995; Paek, 2007). Halenia coreana differs from H. corniculata by having longer, narrower, and incurved spurs and an attenuated leaf apex (Han et al., 2019). Molecular phylogenetic analyses from the nucleotide sequences of the Internal Transcribed Spacer region, and xanthine dehydrogenase (XDH), and chloroplast rbcL indicate that H. coreana forms a distinct clade sister to the clade of populations of H. corniculata from China, Mongolia, Russia, and Japan (Han et al., 2019). The newly established H. coreana is placed under severe risk of extinction due to its extremely small population size and due to human activities. Though it is protected as an endangered species, little is known about the genetic diversity of H. coreana from a conservation perspective. Furthermore, the levels of genetic diversity and differentiation of the narrow endemic H. coreana compared to the widespread H. corniculata have remained unclear.
Halenia coreana is one of three Asian species in Halenia Borkh. It consists of approximately 40 species that are broadly distributed in East Asia and the Americas (Gleason and Cronquist, 1991; Ho and Pringle, 1995; von Hagen and Kadereit, 2003; von Hagen, 2007). The majority of Halenia species are distributed in Mexico and Central America, numbering close to 15 species, and in high alpine regions of the Andes in South America, accounting for approximately 22 species (Allen, 1933; Wilbur, 1984a, 1984b; von Hagen and Kadereit, 2003; von Hagen, 2007). It has been suggested that Halenia originated in East Asia and became dispersed into North America and that it diversified in Central and South Americas (von Hagen and Kadereit, 2003).
Simple sequence repeats (SSR) markers or microsatellites have been commonly used to characterize genetic diversity at the population level in various plant species owing to their co-dominant inheritance, analytical simplicity, and considerable hypervariability (Weber, 1990; Selkoe and Toonen, 2006; Lopez et al., 2015). The next-generation sequence technique facilitated the development of SSR markers (Wang et al., 2018; Yang et al., 2018). For the species of Halenia, SSR markers for H. elliptica D. Don, an Asian species widely used as a traditional medicinal herb in China, were developed (Zhang et al., 2011; Yang et al., 2018). However, genetic diversity in the species of Halenia has not been reported.
In this study, we developed SSR markers for H. coreana and H. corniculata via next-generation sequencing to investigate the genetic diversity of H. coreana. We aim to (1) evaluate the level of genetic diversity in H. coreana, an endangered and endemic species in Korea; (2) to compare the level of genetic diversity with that of H. corniculata, a widespread and sister species of H. coreana; (3) to evaluate the genetic distinctiveness of H. coreana to be recognized as a separate species; and (4) to provide implications pertaining to the conservation biology of H. coreana.
MATERIALS AND METHODSPlant materials
Halenia coreana is known in a few places in Gangwon-do, Jeju-do, and Pyeonganbuk-do (Han et al., 2019). We included 14 individuals found on Mt. Hwaaksan in Gangwon-do. Samples of other populations were not included because fresh materials were not available for analysis. It appears that certain populations, such as the Yongsil population at Mt. Hallasan in Jeju-do, which represents the southern limit of its distributional range, may be extinct, as we failed to locate such plants in repeated fieldwork. Samples of H. corniculata are collected in three populations in China, Japan, and Russia (Table 1). Twenty individuals from each population were included (Table 1). Voucher specimens were deposited in the National Institute Biological Resource (KB).
Selection of SSR markersTotal DNA was isolated from fresh leaves collected in the wild using the DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). To produce high-throughput sequencing data with which to develop SSR markers, we used Mt. Hwaaksan samples. A library was developed using the Illumina MiSeq platform at Macrogen (Seoul, Korea). A total of 68,996,882 raw sequence reads were obtained from the NGS analysis. To achieve loci with low copy numbers, we assembled the filtered reads using Geneious R 10.1.3 (Biomatters Ltd., Auckland, New Zealand) following the method of Cho et al. (2015). We developed 183 primer pairs for microsatellite regions on the sequence set with the primer function in the Geneious program. The forward primers were labeled with 6-FAM or Hex for multiplexing reactions (see Table 2 for the specific fluorescent dye for each locus). To assess the effectiveness of these microsatellite markers, we evaluated the primers for 14 individuals of H. coreana. The initially working primers were further screened using 60 individuals of H. corniculata (Table 1). We then performed polymerase chain reaction (PCR) amplification for validation and genotyping.
The PCR reactions were performed under the following condition: initial denaturation at 95°C for 2 min; 35 cycles of denaturation at 95°C for 20 s, annealing at 58°C for 40 s, and extension at 72°C for 1 min; and a final extension at 72°C for 7 min. The PCR products were analyzed on an ABI 3730XL sequencer with the GeneScan 500 LIZ Size Standard (Thermo Fisher Scientific, Waltham, MA, USA). Allele sizes and peaks for each sample were determined with Peak Scanner Software version 2.0 (Thermo Fisher Scientific).
Genetic analysisBasic genetic properties, i.e., the number of alleles, the expected degree of heterozygosity (HE), and the observed heterozygosity (HO) were calculated using GenAlEx version 6.502 (Peakall and Smouse, 2006, 2012). Deviations from Hardy–Weinberg equilibrium were estimated with GENEPOP version 4.6.9 (Rousset, 2008). The number of alleles, major allele frequency, polymorphism information content, F statistics index (FST), and Nei’s genetic distance were calculated for each population separately. The results of the Mantel test of the association between genetic distance and geographic distance were calculated in GenAlEx. A principal coordinate analysis (PCoA) was conducted to find the genetic clustering using Nei’s genetic distances in GenAlEx.
Genotype data for the SSR markers were analyzed in the model-based STRUCTURE v. 2.3.4 software (Pritchard et al., 2000) to determine the most probable number of clusters (K value) and to assign individuals to different clusters. The K value was determined by running an admixture and a related frequency model with K = 1 to 4 (ten replications per K value); 1,000,000 Markov Chain Monte Carlo iterations were implemented with the burn-in period of each run set to 100,000. The website program STRUCTURE HARVESTER (Earl and vonHoldt, 2012) was used to estimate the optimal K value; this program follows the ΔK method of Evanno et al. (2005).
RESULTSWe developed and selected 17 polymorphic microsatellite loci with clear, strong bands for each allele in the 74 individuals of H. coreana and H. corniculata from the 183 initially designed primer pairs (Table 2). These SSR markers would detect di-, tri-, tetra-, and pentanucleotide repeats (Table 2). In total, 74 alleles were found. The number of alleles (NA) ranged from two (HC71) to ten (HC12), with a mean of 5.0 alleles per locus (Table 2).
The indices of genetic diversity for the sampled populations are summarized in Table 3. The average number of alleles per locus (NA) is slightly smaller in H. coreana at 1.471 than in H. corniculata, where it ranges from 1.765 (China) to 1.882 (Japan). The values of observed heterozygosity (HO) and expected heterozygosity (HE) were lower in H. coreana compared to H. corniculata. The values of HO and HE in H. coreana were 0.038 and 0.119, respectively, in corresponding ranges of 0.035 to 0.112 (mean: 0.075) and 0.178 to 0.262 (mean: 0.227), in H. corniculata (Table 3). The inbreeding coefficient for each locus (FIS), referring to the deviation of the actual frequency of the genotype from the theoretically expected frequency in the population, was 0.0682 in H. coreana and ranged from 0.535 to 0.865 (mean: 0.651) in H. corniculata. The values in both species show no significant differences between the two species on average, but the value of FIS in the Russian population was found to be highest among the sampled populations.
The properties of the diversity indices for the 17 SSR loci are provided in Table 4. Many loci developed in this study showed monomorphism in a population (Table 4). For example, 11 of the 17 loci in H. coreana were monomorphic. Some of the loci, specifically HC03, HC11, H12, HC33, and HC36, were found to be variable during the analyses of the populations of H. corniculata. For the marker HC21, only one allele was found in each of four populations, indicating that this locus is monomorphic for all populations. However, the sizes of the DNA fragments in the different populations varied. These sizes were 350 bp in the Korean population, 271 bp in Japan, 273 bp in China, and 348 bp in Russia. Thus, the locus is polymorphic when all populations are considered.
The pairwise genetic differentiation coefficient (FST) ranged from 0.200 (HC71) to 1 (HC21). The pairwise genetic differentiation coefficient (FST) showed that H. coreana was highly differentiated from H. corniculata (Table 5). A similar tendency was found for Nei’s genetic distance; H. coreana showed relatively high values in the pairs with H. coreana (mean genetic distance, 1.907) (Table 5). The Mantel test using pooled data from the two species showed a weak positive correlation (r = 0.276, p = 0.17) between the geographic distance and the genetic distance (Fig. 1).
To obtain information about the population structures of the H. coreana and H. corniculata accessions from allelic frequencies, we used two methods: STRUCTURE and PCoA. In the STRUCTURE analysis, the computation of Evanno’s ΔK indicated K = 4 as the most likely model (Fig. 2), suggesting the presence of four main groups. With 0.83 as the likelihood (Q-value) to cluster each accession in the seven clusters, a total of 74 individuals were grouped into one of four groups. The China and Russia populations showed a high proportion (Q-value > 0.8). Each population had a low admixture percentage (19%). The grouping determined by STRUCTURE was not related to the geographic distance between the populations. The results of PCoA showed that H. coreana was clearly separated from H. corniculata and that individuals of each population were cohesively clustered (Fig. 3).
DISCUSSIONIt is important to conserve the genetic diversity of the natural population of a species. In particular, this is critical for endangered species with a narrow distribution range and small population size, such as H. coreana. This study is the first to investigate the genetic diversity and population structure of H. coreana and H. corniculata through microsatellite markers.
By genotyping the two species with SSR markers, we show the vulnerability of critically endangered species with a small population size. The genotyping data indicate that H. coreana has low genetic diversity compared to its sister H. corniculata (Table 3). Measures of genetic diversity, such as the percentage of polymorphism, the observed heterozygosity, the expected heterozygosity, and Shannon’s information index, in H. coreana were lower than those the three populations of China, Japan, and Russia of H. corniculata (Table 3).
This pattern may be related to the breeding system, in which high inbreeding coefficient (FIS) values (Rousset, 2002) were found in both H. coreana and H. corniculata (Table 3). The corresponding values in Halenia were much higher than those reported in other species (Lee et al., 2018; Wang, 2020; Wu et al., 2020), suggesting that inbreeding frequently occurs in the two species of Halenia. Kim et al. (2018) found that 56% of flowers in the Mt. Hwaaksan population are cleistogamous. Cleistogamous flowers are very small (3–5 mm in diam. vs. 1.1–1.4 cm in diam. in chasmogamous flowers) having a perianth that remains closed where the stigma is pollinated from the pollen of the same flower, resulting in an inbreeding system. The frequency of cleistogamous flowers in the Mt. Daeamsan population is even higher at 70% (Kim et al., 2018). Such an inbreeding mating system may be advantageous for colonizing a new population, in which a few plants are established or pollinators are not available for the plant. However, not all seeds produced in a population are derived from inbreeding. The flowers of H. coreana and H. corniculata have long spurs in which nectar is produced (von Hagen and Kadereit, 2002), an apparatus facilitating outcrossing. A mixed mating system may have been beneficial to increase the probability of colonizing a new population and to enhance genetic diversity so as to avoid inbreeding depression.
Halenia coreana is genetically differentiated from H. corniculata, as inferred by Wright’s FST (Wright, 1978) and Nei’s genetic distance, showing that the pairwise FST values between the species are higher than those among populations within H. corniculata (Table 5). Interestingly, the FST values in H. corniculata indicate strong genetic differentiation within the species. Each of the populations included in this study is well geographically separated, and population isolation may be responsible for the high values of FST.
The results of PCoA and the STRUCTURE analysis show clear genetic differentiation between H. coreana and H. corniculata and among the populations of H. corniculata (Figs. 2, 3). Our SSR data demonstrate that the Korean population of Halenia is clearly separated from the China + Japan + Russia cluster (Fig. 3), supporting the recognition of H. coreana as a distinct species (Han et al., 2019). The strong geographical structuring of H. coreana and H. corniculata suggests the presence of a barrier to prevent gene flows between species and among different populations. This may explain the weak correlation between the geographical distances and pairwise FST (Fig. 1).
Ecological characteristics should be considered when conservation measures are implemented for H. coreana. Plants of H. coreana require an open habitat to survive and maintain the population (Kim et al., 2018). They grow in open places such as meadows, heliports, and roadsides in alpine areas. Because many of these areas are disturbed by human activities, such as mowing and the construction of trails, habitats for H. coreana have decreased, which can lead to the complete extinction of a population. Changes of vegetation in places where the species occurs, specifically developing forests with trees and shrubs, may have a more direct effect on the survival of the population, as they are not found under crowned habitats with dense taller herbaceous plants or woody plants (Kim et al., 2018). The seedlings of biennial H. coreana would not survive under such shady conditions. The population of the Yongsil trail in Mt. Hallasan, located at an elevation of about 1,500 m, is densely covered by perennial Sasa quelpaertensis Nakai, and plants of H. coreana are no longer found.
Habitat preference to open places is also found in H. corniculata. The species is currently widely distributed in high latitude areas of Japan, Sakhalin, the Kuriles, Kamchatka, China, Siberia, and eastern Europe (Toyokuni and Yamazaki, 1993). Halenia coreana is distributed along the southern boundary of the distributional range of H. corniculata such that the Korean endemic species is more vulnerable to climate changes. Conservation strategies for H. coreana should focus on the preservation of open habitats for the species. Restoration of populations from seeds is also recommended, as the plants produce numerous seeds per plant. With regard to in situ restoration efforts, it is very important to manage the habitat conditions preferred by the species. Many rare and threatened species and highly selfing or clonal species, such as H. coreana, show three important characteristics: a high degree of genetic differentiation among populations, low genetic diversity within populations, and a high degree of inbreeding (Ottewell et al., 2016).
In conclusion, our SSR data on the genetic diversity, breeding system, and genetic differentiation of H. coreana along with the ecological characteristics of the species will be useful to those who plan and implement conservation strategies. The present study highlights the utility of microsatellite markers for assessing and monitoring genetic diversity in an endangered species. The data produced in this study can be used as a baseline for future genetic monitoring and species recovery programs.
ACKNOWLEDGMENTSWe are grateful to Jin-Oh Hyun for providing the Russian samples, Young-Dong Kim and Bo-Yun Kim for information about the sampling sites, and Yun Gyeong Choi and Seung-Hyun Hwang for their help in various stages of this research. We also thank the staff at KB for allowing us to examine their herbarium specimens. This work was supported by research grants from the National Institute of Biological Resources of Korea (NIBR-201703201) and the National Research Foundation of Korea (NRF 2020R1I1A3068464).
Fig. 1.Relationship between geographic distance and genetic distance (FST) for Halenia coreana and H. corniculata. Values were calculated treating the two species as one unit. 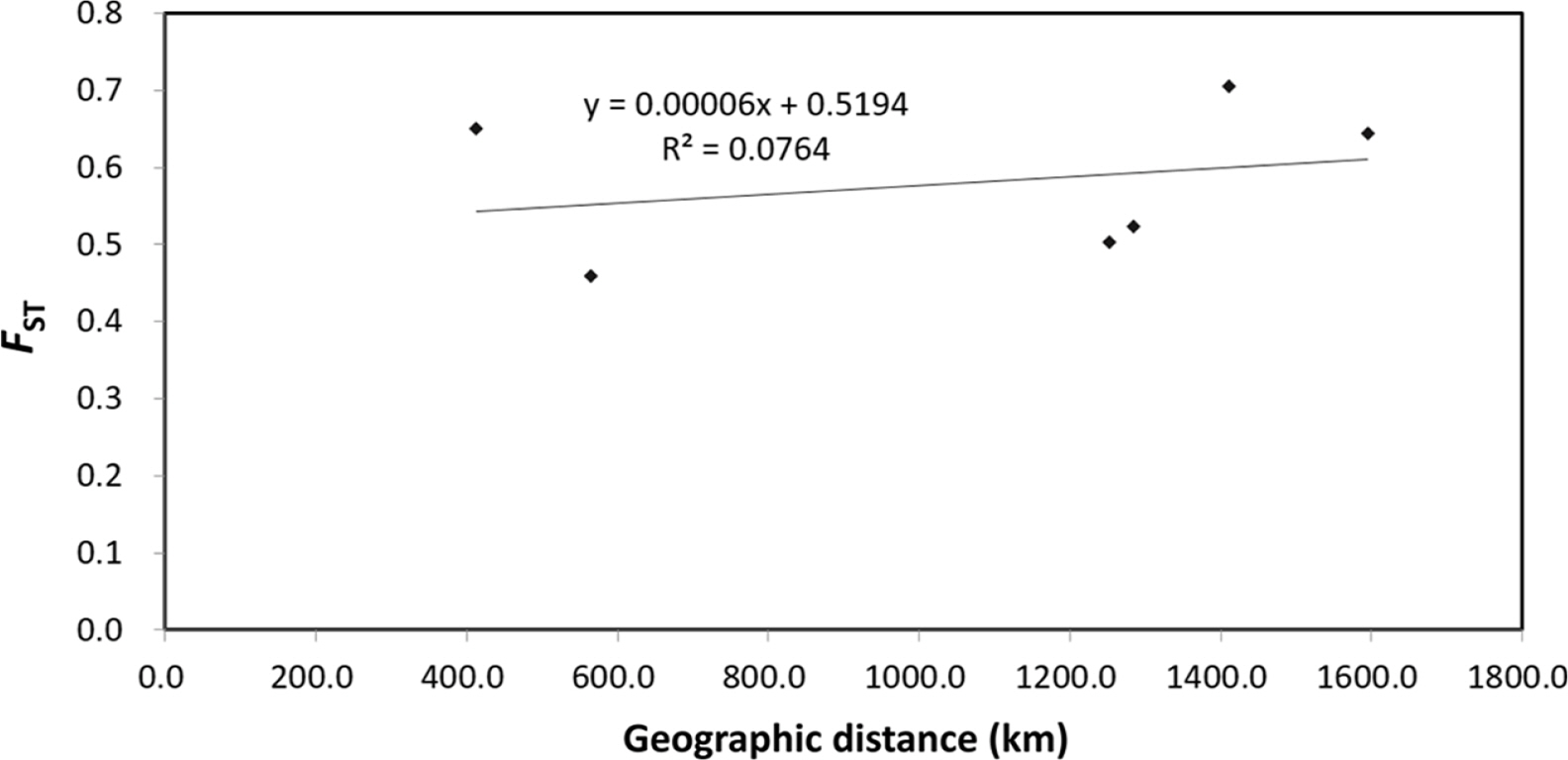
Fig. 2.Results of STRUCTURE analyses based on the simple sequence repeats data and Bayesian model-based clustering analysis for four populations of Halenia coreana and H. corniculata. The bar plot shows the group assignments of 74 individual genotypes for K = 4. 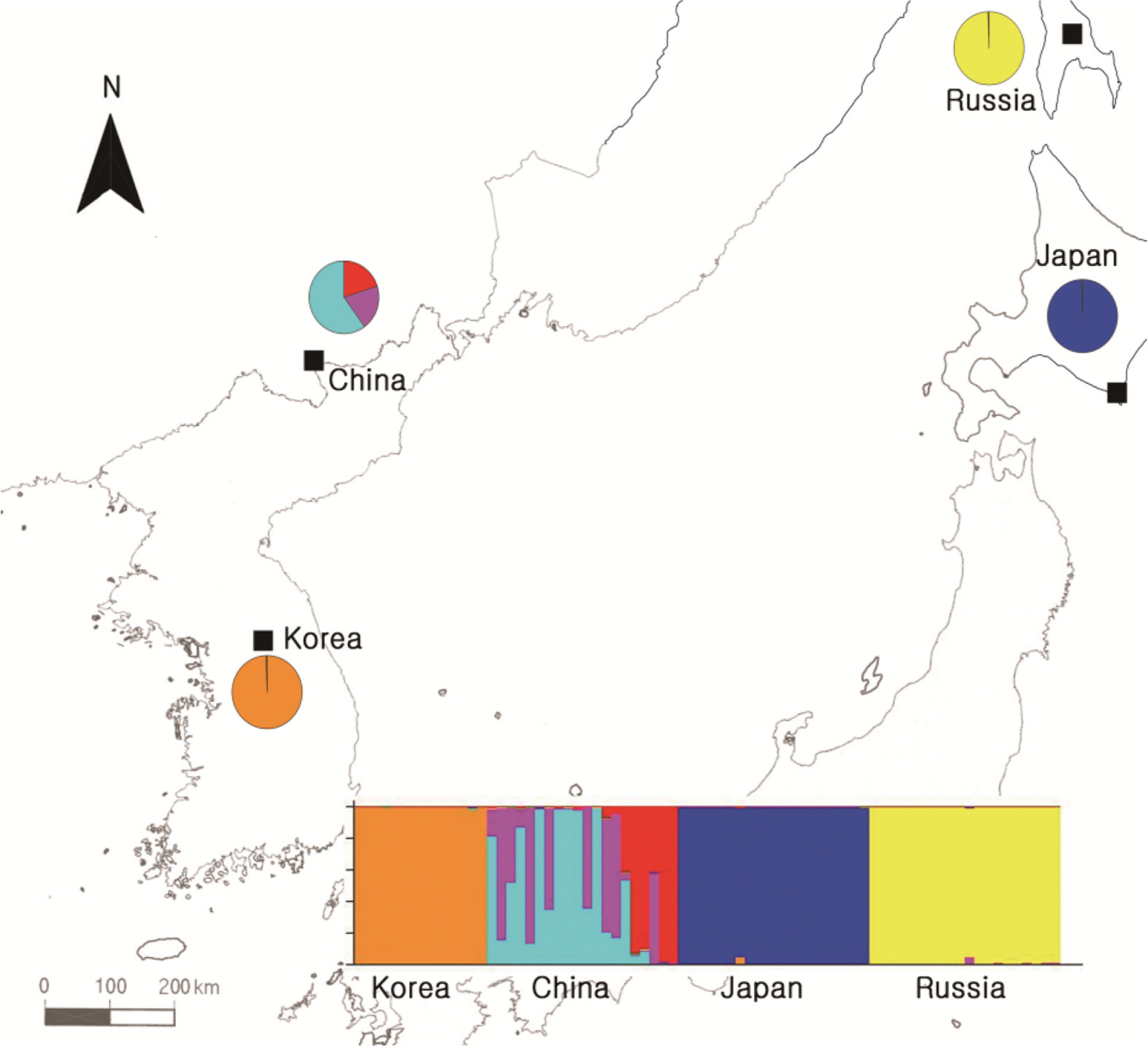
Fig. 3.Principle coordinates analysis of variance based on 17 simple sequence repeats markers. Principal coordinate 1 and 2 account for 30.57% and 23.94% of the variance, respectively. Closed triangle, Halenia coreana; Open triangle, the Chinese population; Closed square, the Russian population; Open square, the Japanese population. 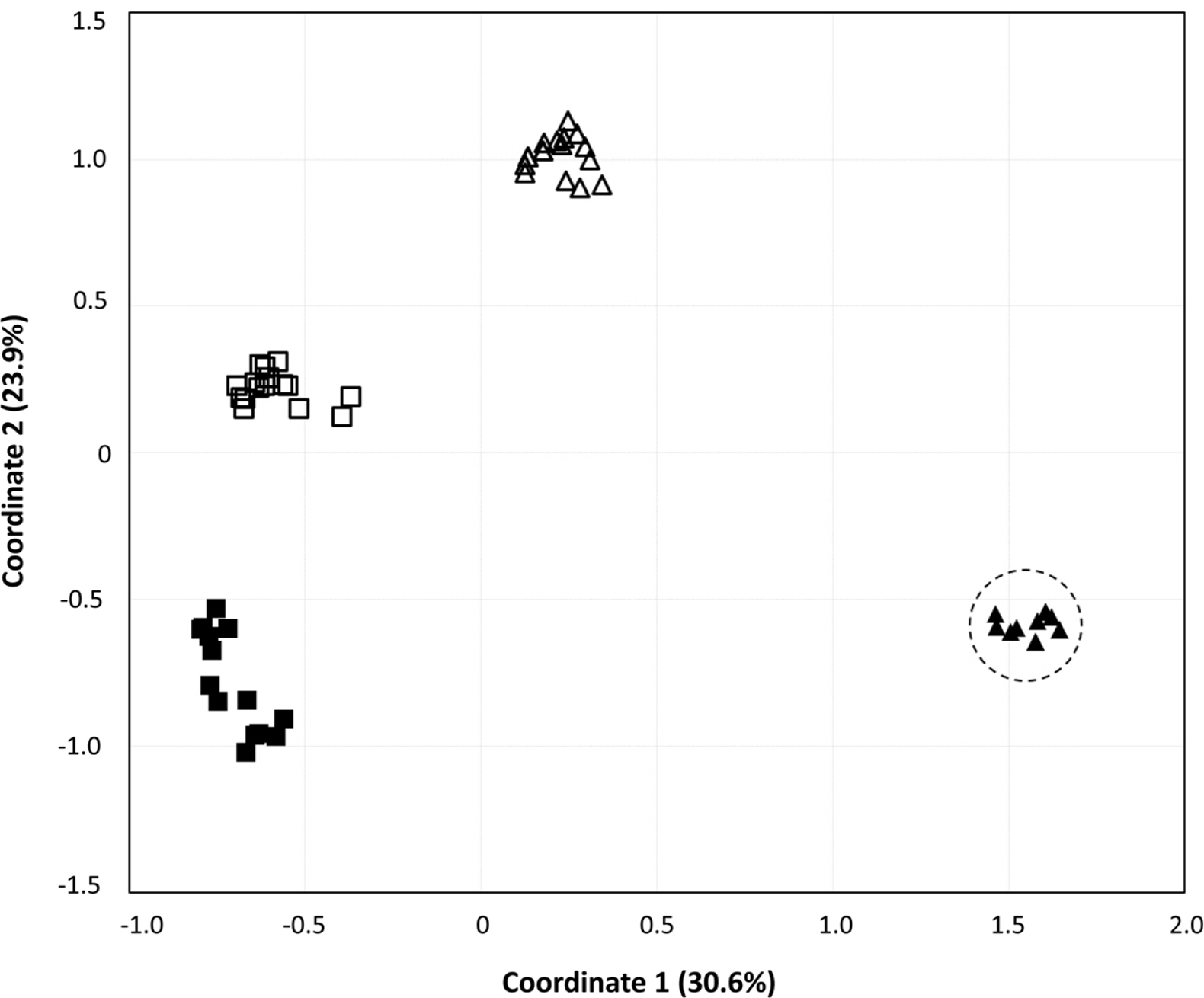
Table 1.Locality and voucher information of the samples of Halenia coreana and H. corniculata used in this study. Table 2.Characteristics of 17 SSR loci developed for Halenia. Table 3.Genetic diversity in Halenia coreana and H. corniculata for geographic population. Table 4.Genetic properties of the 17 polymorphic SSR loci in Halenia coreana and H. coriculata. Table 5.Estimates of pairwise FST (below diagonal) and Nei’s genetic distance (above diagonal) among populations of Halenia coreana and H. corniculata. LITERATURE CITEDAllen, CK. 1933. A monograph of the American species of the genus Halenia
. Annals of the Missouri Botanical Garden 20: 119-222.
Cheek, M. Lughadha, EN. Kirk, P. Lindon, H. Carretero, J. Looney, B. Douglas, B. Haelewaters, D. Gaya, E. Llewellyn, T. Ainsworth, AM. Gafforov, Y. Hyde, K. Crous, P. Hughes, M. Walker, BE. Forzza, RC. Wong, KM and Niskanen, T. 2020. New scientific discoveries: Plants and fungi. Plants People Planet 2: 371-388.
Cho, W-B. Choi, I-S and Choi, B-H. 2015. Development of microsatellite markers for the endangered Pedicularis ishidoyana (Orobanchaceae) using next-generation sequencing. Applications in Plant Sciences 3: 1500083.
Choi, H-J. Yang, S. Yang, J-C and Friesen, N. 2019.
Allium ulleungense (Amaryllidaceae), a new species endemic to Ulleungdo Island, Korea. Korean Journal of Plant Taxonomy 49: 294-299.
Chung, J-M. Shin, J-K. Sun, E-M and Kim, H-W. 2017. A new species of Epilobium (Onagraceae) from Ulleungdo Island, Korea, Epilobium ulleungensis
. Korean Journal of Plant Taxonomy 47: 100-105.
Do, DN. Luong, DV. Nguyen, CD. Hoang, ST. Le, HT. Han, JE and Park, H-S. 2019. A new yellow Camellia (Theaceae) from central Vietnam. Korean Journal of Plant Taxonomy 49: 90-95.
Donoghue, MJ and Alverson, WS. 2000. A new age of discovery. Annals of the Missouri Botanical Garden 87: 110-126.
Earl, DA and vonHoldt, BM. 2012. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4: 359-361.
Evanno, G. Regnaut, S and Goudet, J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Molecular Ecology 14: 2611-2620.
Gleason, HA and Cronquist, A. 1991. Manual of Vascular Plants of Northeastern United States and Adjacent Canada. The New York Botanical Garden, New York. 910.
Han, S.-M. Won, H and Lim, CE. 2019.
Halenia coreana (Gentianaceae), a new species from Korea: Evidence from morphological and molecular data. Phytotaxa 403: 86-98.
Ho, TN and Pringle, JS. 1995. Gentianaceae. In Flora of China. 16: Gentianaceae through Boraginaceae. Wu, ZY. Raven, PH (eds.), Science Press, Beijing and Missouri Botanical Garden Press, St Louis, MO. 1-139.
Kang, D.-H. Ong, HG. Lee, J-H. Jung, E-K. Kyaw, N.-O. Fan, Q and Kim, Y-D. 2021. A new broad-leaved species of loquat from eastern Myanmar and its phylogenetic affinity in the genus Eriobotrya (Rosaceae). Phytotaxa 482: 279-290.
Kim, B-Y. Won, H. Phourin, C. Lim, C-K. Shin, J-S. Kim, Y-S and Cho, S-H. 2019.
Impatiens cardamomensis (Balsaminaceae), a new species from Cambodia. Korean Journal of Plant Taxonomy 49: 319-323.
Kim, KC and Byrne, LB. 2006. Biodiversity loss and the taxonomic bottleneck: Emerging biodiversity science. Ecological Research 21: 794-810.
Kim, YC. Chae, HH. Nam, GH and Lee, KS. 2018. Adapting strategy of biennial endangered Halenia corniculata (L.) Cornaz to environmental stress and its implications for conservation plan. Journal of Plant Biology 61: 177-185.
Lee, S-R. Choi, J-E. Lee, B-Y. Yu, J-N and Lim, CE. 2018. Genetic diversity and structure of an endangered medicinal herb: Implications for conservation. AoB PLANTS 10: ply021.
Lopez, L. Barreiro, R. Fischer, M and Koch, MA. 2015. Mining microsatellite markers from public expressed sequence taqs databases for the study of threatened plants. BMC Genomics 16: 781.
National Institute of Biological Resources. 2012. Red Data Book of Endangered Vascular Plants in Korea. National Institute of Biological Resources, Incheon. 392 pp.
Nam, GH. Jang, H-D. Lee, B-Y and Chung, GY. 2020.
Carex brevispicula (Cyperaceae), a new species from Korea. Korean Journal of Plant Taxononmy 50: 395-402.
Ottewell, KM. Bickerton, DC. Byrne, M and Lowe, AJ. 2016. Bridging the gap: A genetic assessment framework for population-level threatened plant conservation prioritization and decision-making. Diversity and Distributions 22: 174-188.
Paek, W-K. 2007. Gentianaceae. The Genera of Vascular Plants of Korea. Park, C-W (ed.), Academy Publishing Co, Seoul. 764-772.
Peakall, R and Smouse, PE. 2006. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes 28: 288-295.
Peakall, R and Smouse, PE. 2012. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research–an update. Bioinformatics 28: 2537-2539.
Pritchard, JK. Stephens, M and Donnelly, P. 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945-959.
Rousset, F. 2002. Inbreeding and relatedness coefficients: What do they measure? Heredity 88: 371-380.
Rousset, F. 2008. genepop’007: A complete re-implementation of the genepop software for Windows and Linux. Molecular Ecology Resources 8: 103-106.
Selkoe, KA and Toonen, RJ. 2006. Microsatellites for ecologists: A practical guide to using and evaluating microsatellite markers. Ecology Letters 9: 615-629.
Sobral, M and Stehmann, JR. 2009. An analysis of new angiosperm species discoveries in Brazil (1990–2006). Taxon 58: 227-232.
Toyokuni, H and Yamazaki, T. 1993.
Halenia
. Flora of Japan. IIIa: Iwatsuki, K. Yamazaki, T. Boufford, DE. Ohba, H (eds.), Kodansha, Tokyo. 140 pp.
von Hagen, KB. 2007. Description of new taxa of Halenia Borkh. (Gentianaceae) from Colombia and Venezuela with significance for testing a key innovation hypothesis. Organisms Diversity and Evolution 7: 1-11.
von Hagen, KB and Kadereit, JW. 2002. Phylogeny and flower evolution of the Swertiinae (Gentianaceae-Gentianeae): Homoplasy and the principle of variable proportions. Systematic Botany 27: 548-572.
von Hagen, KB and Kadereit, JW. 2003. The diversification of Halenia (Gentianaceae): Ecological opportunity versus key innovation. Evolution 57: 2507-2518.
Wang, H. Yang, H. Liu, P-L. Su, C. Xiao, L and Chang, Z-Y. 2018. Isolation and characterization of microsatellite loci from Oxytropis diversifolia (Fabaceae). Applications in Plant Sciences 6: e01168.
Wang, S-Q. 2020. Genetic diversity and population structure of the endangered species Paeonia decomposita endemic to China and implications for its conservation. BMC Plant Biology 20: 510.
Wilbur, RL. 1984a. A synopsis of the genus Halenia (Gentianaceae) in Central America. Bulletin of the Torrey Botanical Club 111: 366-374.
Wilbur, RL. 1984b. A synopsis of the genus Halenia (Gentianaceae) in Mexico. Rhodora 86: 311-337.
Wright, S. 1978. Evolution and the Genetics of Populations. 4: Variability within and among Natural Populations. University of Chicago Press, Chicago. 590 pp.
Wu, Q. Zang, F. Ma, Y. Zheng, Y and Zang, D. 2020. Analysis of genetic diversity and population structure in endangered Populus wulianensis based on 18 newly developed EST-SSR markers. Global Ecology and Conservation 24: e01329.
Yang, M. Han, N. Li, H and Meng, L. 2018. Transcriptome analysis and microsatellite markers development of a traditional Chinese medicinal herb Halenia elliptica D. Don (Gentianaceae). Evolutionary Bioinformatics 14: 1-6.
|
|
|||||||||||||||||||||||||||||||||||||||