AbstractNeutral genetic diversity found in plant species usually leaves an indelible footprint of historical events. Korea’s main mountain range (referred to as the Baekdudaegan [BDDG]), is known to have served as a glacial refugium primarily for the boreal and temperate flora of northeastern Asia. In addition, life-history traits (life forms, geographic range, and breeding systems) influence the within- and among-population genetic diversity of seed plant species. For example, selfing species harbor significantly less within-population genetic variation than that of predominantly outcrossers. A previous study of two Liparis species (L. makinoana and L. kumokiri) emphasizes the role of the abovementioned factors shaping the levels of genetic diversity. Liparis makinoana, mainly occurring on the BDDG and self-incompatible, harbors high levels of within-population genetic diversity (expected heterozygosity, HeP = 0.319), whereas there is no allozyme variation (HeP = 0.000) in L. kumokiri, which is self-compatible and mainly occurs in lowland hilly areas. To determine if this trend is also found in other congeners, we sampled five populations of L. krameri from the southern part of the Korean Peninsula and investigated the allozyme-based genetic diversity at 15 putative loci. The somewhat intermediate levels of within-population genetic variation (HeP = 0.145) found in L. krameri are most likely due to its occurrence in mountainous areas that, despite being outside of the main ridge of the BDDG, still served as refugia, and a self-incompatible breeding system. Management strategies are suggested for L. krameri and L. makinoana based on the levels and distribution of genetic diversity and inbreeding.
Neutral genetic markers (amplified fragment length polymorphisms, allozymes, inter simple sequence repeats, microsatellites, etc.) have been successfully adopted to study levels of genetic diversity (Hamrick and Godt, 1989; Nybom, 2004), genetic structure (Hamrick et al., 1989; Schnabel et al., 1998) including fine-scale spatial genetic structure (Vekemans and Hardy, 2004; Chung et al., 2007; Gonzales et al., 2010), mating systems (Ritland and Jain, 1981; Whitehead et al., 2018), seed dispersal patterns and gene flow (Grivet et al., 2005; Burczyk et al., 2006; Troupin et al., 2006; Jordano et al., 2007), and parentage analyses (Hardesty et al., 2006; Sezen et al., 2009), among others. Single gene (or genetic) markers (nuclear, chloroplast DNA, and less frequently mitochondrial DNA) are particularly useful in documenting levels of between-population historical seed flow and reconstructing the biogeographical history of plant species (i.e., phylogeography). Therefore, historical events (e.g., glaciations) could leave a ‘non-negligible’ trace on the levels of neutral genetic diversity found within species.
Some life-history and ecological traits of seed plants are directly or indirectly related to dispersal of genes (i.e., pollen and seed flow), inbreeding, and effective population sizes (Ne) (i.e., mating systems, etc.) which, in turn, have causal relationships with the levels and the distribution of genetic diversity (Loveless and Hamrick, 1984; Hamrick and Godt, 1989, 1996). Plant-allozyme literature (Hamrick and Godt, 1989; Olson et al., 2016) demonstrate that outcrossing species or those with a mixed mating system harbor more genetic diversity than selfing species, which is generally true both when heterospecific (mixed species) and when congeneric comparisons are carried out. Furthermore, outcrossing or mixed mating species usually show a lower degree of between-population genetic differentiation than selfing species.
On the Korean Peninsula the Baekdudaegan (BDDG) (Fig. 1) is regarded as a sort of ‘backbone’ because it stretches across the peninsula with over 1,600 km. The BDDG is a biodiversity hotspot primarily due to its role as a Pleistocene refugium (Borzée et al., 2017; Chung et al., 2017, 2018a). We found a pattern of high within-population (expected heterozygosity, mean HeP = 0.159) and low to moderate between-population allozyme-based genetic differentiation (mean GST = 0.175) in 16 plant species centered in the BDDG (Chung et al., 2017). Such a pattern of genetic “health” for plant populations growing in these mountains would be ascribed to the likely large Ne and great demographic stability during the Last Glacial Maximum.
Considering these two factors (i.e., historical events and current mating systems), one may expect that outcrossing plant species that dwelled on the BDDG for a long period of time would maintain higher within-population genetic variation and lower between-population genetic divergence than predominantly selfing species that occur in lowland hills. An example that fits this model is the terrestrial orchid congeneric pair Liparis makinoana Schltr. and L. kumokiri F. Maek. in South Korea (Chung et al., 2007). Consistent to the expectation, L. makinoana, mainly occurring on the BDDG and self-incompatible, harbors high levels of within-population genetic diversity (HeP = 0.319; FST = 0.107), whereas there is no allozyme variation (HeP = 0.000) in L. kumokiri, largely occurring in lowland hills and self-compatible (Chung et al., 2007).
In order to determine if this trend is also found in other congeners of Liparis, we selected the congener L. krameri Franch. & Sav. from southern Korean Peninsula as a study species. Liparis makinoana is relatively continuously distributed along the BDDG, whereas most populations of the self-incompatible L. krameri occur in mountainous locations away from the BDDG and are relatively discontinuously distributed. Given these traits, we expect a lower within-population genetic variation and a higher between-population genetic differentiation for L. krameri compared to L. makinoana. In addition, we determined the genetic type to which this species belongs, based on the observed values for the three main genetic parameters (GST, HeP, and FIS) as described in Ottewell et al. (2016). Finally, we suggested appropriate management strategies for the three Liparis species.
Materials and MethodsStudy species, population sampling, and allozyme electrophoresisThe perennial terrestrial orchid Liparis krameri is distributed in mainland China (SW Hubei), the Korean Peninsula, Russian Far East, and Japan (Chen et al., 2009). On the Korean Peninsula, the species grows well on half-shaded slopes under trees (e.g., pine-oak forests) rich in humus with a good drainage. Pollinators of L. krameri are unknown, but the species is visited by small diptera such as fungus gnat in Japan (K. Suetsugu, pers. comm.). In LMR-4, ovaries from induced autogamy were detached, but those pollinated from different individuals set fruit, suggesting self-incompatibility. However, in Japan L. krameri is either self-compatible (but non-autogamous) or self-incompatible (thus, ‘partially self-incompatible’); fruit and seed set of selfed individuals was lower than that of crossed ones (K. Suetsugu, pers. comm.). A similar scenario is found in L. makinoana: it was self-incompatible based on individuals in a location near Mt. Sobaek, South Korea (Oh et al., 2001). However, as L. makinoana in Japan set fruits with viable seeds sometimes produced by induced autogamy, Suetsugu considered it as ‘partially self-incompatible’ (K. Suetsugu, pers. comm.). Whitehead et al. (2018) showed that plant mating systems often vary among conspecific populations based on meta-analyses of 741 populations from 105 species and cautions that estimates of outcrossing rates from single populations are often unreliable indicators of the entire species.
To investigate allozyme variation within- and between-populations of L. krameri, we collected leaf samples (from a total of 177 individuals) from five populations from southern Korean Peninsula (Fig. 1, Table 1). We cut 1 cm from the leaf tip to minimize the damage to plants; we transported leaf samples on an ice box and kept them in a refrigerator once at the corresponding author’s laboratory. We cut and crushed them with a precooled mortar and pestle in a phosphate polyvinylpyrrolidone extraction buffer (Mitton et al., 1979). We absorbed enzyme extracts onto 4 × 6-mm wicks cut from Whatman 3 MM chromatography paper (Whatman International, Maidstone, UK), which were then stored at −70°C until needed.
To compare L. krameri with the previously studied (in the same laboratory) L. kumokiri and L. makinoana (Chung et al., 2005, 2007), we followed the same method as described by Chung et al. (2007). We stained starch gels (12%) for 15 putative loci resolved from nine enzyme systems using three buffer systems: Dia-1, Dia-2, Fdh, Idh-1, Idh-2, Lap-1, Lap-2, Mdh-1, Mdh-2, 6Pgd-1, 6Pgd-2, Pgi-1, Pgi-2, Pgm, and Skdh. We designated putative loci sequentially, with the most anodally migrating isozyme designated as ‘1,’ the next ‘2,’ and so on. We also designated different alleles within each locus sequentially by alphabetical order (‘a’, ‘b’, ‘c’, ‘d’, and ‘e’).
Data analysisWe designated a locus as ‘polymorphic’ when two or more alleles were detected, regardless of their frequencies. We estimated the genetic diversity parameters within populations using the programs POPGENE (Yeh et al., 1999) and FSTAT (Goudet, 1995): percentage of polymorphic loci (%PP), mean number of alleles per locus (AP), allelic richness (AR) using a rarefaction method to account for uneven population sample sizes (Hurlbert, 1971; El Mousadik and Petit, 1996), observed heterozygosity (HoP) and Hardy–Weinberg (H–W) expected heterozygosity or Nei’s (1978) gene diversity (HeP). Except for AR and Ho, we also estimated these parameters for the total samples as a whole (i.e., at the species level). Hereafter, the subscript “P” indicates population means, while the subscript “S” indicates species’ (or pooled samples) means.
To test for differences between populations of L. makinoana and those of L. krameri for observed statistics, OSx (ARP, HoP, and HeP), we used a permutation scheme (999 replicates) by randomly allocating whole samples to the different groups, keeping the number of samples in each group constant and calculating differences between populations of the two species for randomized statistics, RSx. We then obtained the p-value of the test as the proportion of randomized data sets giving higher values for RSx than for OSx. These calculations were performed using FSTAT (Goudet, 1995).
To test for recent decreases of NeP (i.e., genetic bottlenecks) in L. krameri, we evaluated differences across loci between the H–W He and the equilibrium heterozygosity (Heq) expected assuming mutation–drift equilibrium. Using the program BOTTLENECK (Cornuet and Luikart, 1996), we evaluated these differences using a sign test and a Wilcoxon sign-rank test under an infinite allele model. Since allelic diversity is generally lost more rapidly than HeP (Nei et al., 1975), recently bottlenecked populations will exhibit an excess of H–W HeP relative to Heq (Cornuet and Luikart, 1996; Luikart et al., 1998).
We used the program SPAGeDi (Hardy and Vekemans, 2002) to calculate population-level FIS (inbreeding) and its significance level by 999 permutations under the null hypothesis of FIS = 0. To measure deviations from H–W equilibrium at each polymorphic locus, we calculated averages of Wright’s (1965) FIS and FST (deviations from H–W equilibrium of individuals relative to their local populations, and local populations relative to the total population, respectively) following Weir and Cockerham (1984). Using FSTAT, we constructed 95% bootstrap confidence intervals (CI) (999 replicates) around means of FIS and FST, and considered the observed FIS and FST to be significant when 95% CI did not overlap zero. Statistical significance of differences in FIS and FST between populations of L. makinoana and L. krameri was determined as outlined above for AR, HoP, and HeP.
To estimate the levels of genetic divergence among populations, we performed a principal coordinate analysis (PCoA) with GenAlEx (Peakall and Smouse, 2012) based on codominant genotypic distances. In addition, we assessed the genetic structure by means of the Bayesian algorithm implemented in STRUCTURE 2.3.4 (Pritchard et al., 2000). The program estimates the likelihood of the individuals being structured in a given number of genetic clusters (or genetic populations, K). We selected ‘admixture’ and ‘correlated’ as appropriate models for ancestry and allele frequencies, respectively, as events of migration and populations with shared ancestry are presumably to occur in L. krameri. We set the burn-in period and Markov Chain Monte Carlo to 50,000 and 500,000 iterations, respectively, and 20 replicates per K were run. We determined the most likely value of K by the ΔK statistics of Evanno et al. (2005). As the ΔK method tends to identify K = 2 as the top level of hierarchical structure (Janes et al., 2017), we combined it with the method of choosing the smallest K after the log probability of data [ln Pr(X|K)] values reached a plateau (Pritchard et al., 2010). The results of both methods were visualized with the aid of Structure Harvester (Earl and vonHoldt, 2012).
ResultsLevels of genetic variation within populations and samples as a wholeSeven (Dia-1, Dia-2, Pgm, Idh-1, Pgi-2, 6Pgd-1, and 6Pgd-2) out of 15 loci were polymorphic across five populations of L. krameri in southern Korean Peninsula. Variation at allozyme loci within populations was moderate or high (Table 1). At the population level, the average percentage of polymorphic loci (%PP) was 41.1%, the mean number of alleles per locus (AP) was 1.59, the AR was 1.55, and the mean observed (HoP) and expected heterozygosity (HeP) were 0.136 and 0.145, respectively (Table 1). Higher levels of %PS (46.7%), AS (1.93), and HeS (0.166) were found when samples were treated as a whole (Table 1). Liparis makinoana harbored higher within-population genetic variation than L. krameri (one-sided p-values for ARP, HoP, and HeP were 0.004, 0.010, and 0.010, respectively).
Among the five studied populations of L. krameri, we did not find any population with significant excess of H–W He under the infinite allele model (Table 2).
Population genetic structureWe found no significant deficiency of heterozygotes (at the 0.05 level) relative to H–W expectations in all but one population (LMR-4; FIS = 0.186) (Table 2). These results, as well as the non-significant multi-population-level FIS (FIS = 0.059; 95% CI, −0.047 to 0.146) (Table 2), indicated a general fit to H–W expectations within populations. The value of pooled multi-population FIS for L. makinoana (FIS = 0.199) was significantly larger than that of L. krameri (p = 0.025). Deviation from H–W expectations due to allele frequency differences between populations was significantly different from zero for the two species (FST = 0.149; 95% CI, 0.095 to 0.216 for L. krameri vs. FST = 0.107; 95% CI, 0.067 to 0.148 for L. makinoana). We found no statistically significant differences between the two species (p = 0.587).
In the PCoA (Fig. 2), the first two components accounted for 78.1% (axis 1 = 45.0%; axis 2 = 33.1%) of the total genetic variance. Population LMR-3, which occurs in Gajisan Mt. and is located very close to LMR-1 and LMR-2, clustered, instead, with LMR-4 (Fig. 2). The best clustering scheme of STRUCTURE (K = 2 or K = 3, according to both the ln Pr(X |K) and the ΔK statistic), agreed with the PCoA (Fig. 3).
DiscussionLevels and distribution of genetic diversity of L. krameriWe anticipated a lower within-population genetic variation and a higher between-population genetic differentiation for L. krameri compared to L. makinoana. Our results support the expectation for within-population genetic variation, but not that for between-population genetic differentiation; although, as predicted, FST for L. makinoana was lower than that for L. krameri (0.107 vs. 0.149), these two values were not statistically significantly different.
The levels of genetic variation within populations of L. krameri and within the species (i.e., the samples pooled as a whole) are slightly lower than those compiled for plant species occurring mainly in the BDDG (HeP = 0.145 vs. 0.159 and HeS = 0.166 vs. 0.193) (Table 1). However, L. krameri maintains higher levels of genetic variation at population and species levels than those typically found in orchids (HeP = 0.134 and HeS = 0.135) (Table 1), in plant species with outcrossing-animal breeding system (HeP = 0.124 and HeS = 0.167) (Table 1), and in herbaceous perennials (HeP = 0.096 and HeS = 0.125) (Table 1).
Except LMR-4, HoP is close to HeP (multi-population-level FIS = 0.059), which in part is consistent with the life-history traits observed for the species in Korea (self-incompatibility and pollination by unidentified insects). However, a significant deficiency of heterozygotes relative to H–W expectations (multi-population-level FIS = 0.199) was found in L. makinoana.
Liparis krameri exhibits a relatively low degree of genetic differentiation among populations (FST = 0.149). This degree is lower than those compiled for plant species occurring mainly in the BDDG (GST = 0.175) (Table 1), orchid species (GST = 0.190) (Table 1), plant species with outcrossing-animal breeding system (GST = 0.197) (Table 1), and perennial herbs (GST = 0.256) (Table 1). Why do populations of L. krameri harbor moderate to high levels of within-population genetic variation and relatively low degree of between-population genetic divergence? Although most populations of L. krameri occur in mountainous locations away from the BDDG (which has been suggested to be one of the main East Asia refugia for plants) (Chung et al., 2017), the species would have endured the glacial periods in favorable areas for the survival of the species. In addition to the BDDG, some of its ‘branches’ or associated mountain systems, such as ‘Nakdongjeongmaek’ (where Gajisan Mt. is located, home of populations LMR-1 to LMR-3) (Fig. 1), as well as Jeju Island (where LMR-5 is located) (Fig. 1), would have also acted as suitable refugia for many temperate and boreal plants; populations within refugia would have maintained relatively large Ne and connectivity among them (Dolezal et al., 2012; Chung et al., 2013, 2017). Orchids produce tiny seeds that have the potential for long distance dispersal by wind or storms (Arditti and Ghani, 2000; Trapnell and Hamrick, 2004; Yukawa et al., 2012; Takashima et al., 2016). As L. krameri occurs in southern Japan, one may consider that current Korean populations might have originated by postglacial expansion from outside of the peninsula. This scenario is, however, less likely than in situ survival in glacial refugia because L. krameri harbors high levels of HeP. Analysis of ecological niche modeling would provide us insights into Pleistocene demographic history of L. krameri on the Korean Peninsula. However, given the limited number of presence records in South Korea and little information on the distribution of L. krameri in North Korea, it is unlikely that niche modeling will generate accurate and useful predictions.
Liparis krameri is genetically healthyAccording to population genetics theory, the “genetic health” of populations is characterized by large Ne, sufficient gene flow to counteract the effects of random genetic drift, and a low degree of inbreeding (Ottewell et al., 2016). These factors are closely related to the three major population genetic parameters GST, HeP, and FIS: in healthy populations or species, one would expect low degrees of GST and low levels of FIS but high levels of HeP. Ottewell et al. (2016) developed a plain language genetic assessment approach by setting eight possible combinations based on the rating of the three parameters as ‘low’ (L) or ‘high’ (H).
The FST value found in L. krameri (0.149) can be considered as ‘low’ because it is lower than the threshold value (FST = 0.150) proposed by Ottewell et al. (2016), but also because it is lower than the average values reported for plant species occurring mainly in the BDDG, orchid species, plant species with outcrossing-animal breeding system, and perennial herbs (Table 1). The value of HeP (0.145) can be considered as ‘high’ because, despite being lower than the value found in L. makinoana (HeP = 0.319) (Table 1), it is higher than the average values reported for orchids, plant species with outcrossing-animal breeding system, and herbaceous perennials (Table 1). Finally, we rate the third parameter FIS (0.059) as ‘low’ because it does not deviate from the null hypothesis of FIS = 0 and it is lower than its congener L. makinoana (FIS = 0.199) (Table 1) and than what is expected for herbaceous perennials (FIS = 0.174) (Duminil et al., 2009). Thus, in terms of all (pooled) populations examined, L. krameri in southern Korean Peninsula can be represented as L/H/L (which can be regarded as a “healthy” species). According to table 1 in Ottewell et al. (2016), L/H/L belongs to the first genetic management strategy which is characterized by the following species’ ecological and demographical scenarios: (1) historically connected populations maintaining gene flow contemporarily; (2) reduced effects of outbreeding depression and local adaptation because of somewhat homogeneous habitats (the species grows well on half-shaded slopes under trees forests rich in humus with a good drainage); (3) populations acting as “source/sink” metapopulations; and (4) avoidance of loss of genetic diversity and inbreeding due to large Ne. On the basis of these scenarios, Ottewell et al. (2016) recommend focusing on maintaining and facilitating metapopulation dynamics, although translocations between populations to boost population sizes is also a viable option.
In terms of individual populations, the only way to know whether a population is low (L) or high (H) regarding FST is by getting a matrix of pairwise FST values and check whether a given population has low or high levels of FST with the rest of populations. There were no significant differences among the five means (from 0.107 in LMR-2 to 0.154 in LMR-5) for FST pairwise values of each population with the rest of the populations (Kruskal-Wallis test, H = 5.287, p = 0.259). Thus, we considered that the allele frequencies are rather homogenous among the five populations and rated each FST mean as ‘L’ because Wright’s FST (0.149) estimated from five populations was considered as ‘L’. Except LMR-4, all populations of L. krameri could be expressed as L/H/L, and, thus, could be considered as “healthy” ones. The population LMR-4 could be represented as L/H/H because FIS (0.186) is similar to that of L. makinoana (FIS = 0.199) and slightly higher than the mean reported for herbaceous perennials (FIS = 0.174) (Duminil et al., 2009). According to the ecological/demographical scenarios proposed by Ottewell et al. (2016), recent fragmentation or reduction in population size would have occurred in LMR-4, causing inbreeding. As management strategies, approaches to reduce inbreeding (e.g., facilitation of pollen/seed immigration and active translocations) should be initiated (Ottewell et al., 2016).
Finally, as L. makinoana could be represented as L/H/H, for the long-term survival of this species management strategies should be conducted as suggested above for LMR-4. As L. kumokiri is a genetically depauperate species at neutral loci and mainly occurs under forests showing a narrow ecological amplitude, it might lack adaptability to environmental changes and, thus, is prone to extinction. The species blooms during the rainy season and is rain-triggered self-pollinated (Suetsugu, 2019). From a conservation and restoration perspective, L. kumokiri is of higher priority regarding conservation and restoration actions, which should be focused on increasing the number of individuals and populations.
ACKNOWLEDGMENTSThe authors thank Beom Jin Shim and Myeong Soon Park for laboratory assistances. This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2013R1A1A2063524 to MYC and NRF-2013R1A1A3010892 and NRF-2017R1A2B4012215 to MGC) and was carried out as part of the Infrastructure for the Conservation and Restoration of Rare and Endemic Plants in Korea National Arboretum that supported to MGC from 2015 to 2019.
NOTES
ORCID: Mi Yoon CHUNG https://orcid.org/0000-0002-8756-5367; Kangshan MAO https://orcid.org/0000-0002-0071-1844; Jordi LÓPEZ-PUJOL https://orcid.org/0000-0002-2091-6222; Myong Gi CHUNG https://orcid.org/0000-0002-1283-3574
Fig. 1.Locations of sampled populations of by the five populations of Liparis krameri separated by in South Korea. LMR-1 to LMR-3 are located at Gajisan Mt. LMR-1 and LMR-2 is separated by ca. 750 m; LMR-2 and LMR-3 are separated by ca. 560 m. Population LMR-3 is located near the peak at 1,240 m. LMR-4 (elev. 440 m) and LMR-5 (elev. 690 m) were from Geojedo Island and Jejudo Island, respectively. The thick line indicates the main ridgeline of the Baekdudaegan (BDDG) mountain system, and the dotted line represents the so-called “Nakdongjeongmaek,” one of the 13 mountainous branches of the BDDG. 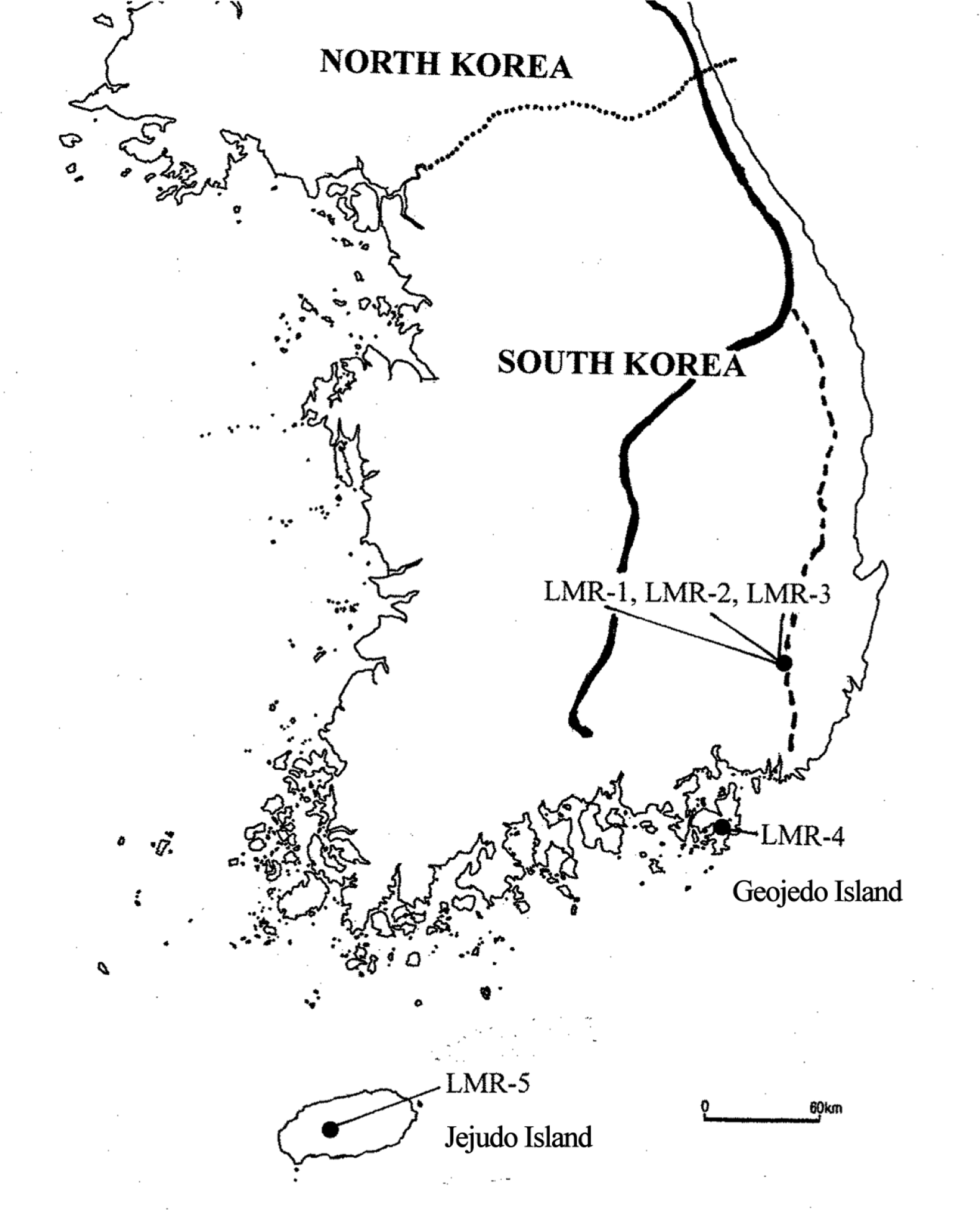
Fig. 2.The principal coordinate analysis (PCoA) of the 5 studied populations of Liparis krameri in South Korea. Blue circles, populations on Gajisan Mt. (LMR-1 to LMR-3); green circle, population on Geoje Island (LMR-4); yellow circle, population on Jeju Island (LMR-5). 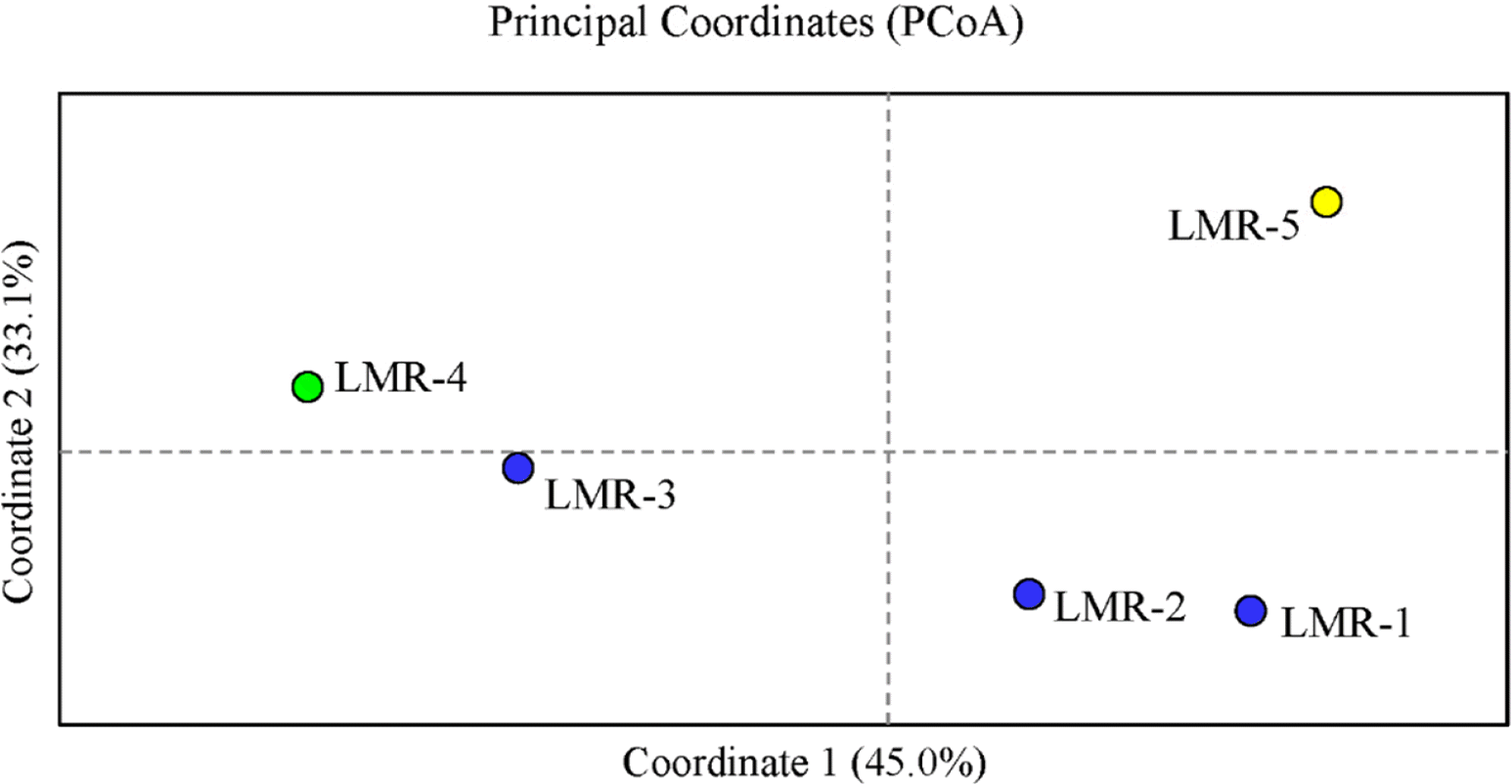
Fig. 3.Results of STRUCTURE analysis for all studied individuals of the five populations of Liparis krameri in South Korea. A, B. The most likely K was estimated by the ΔK statistics (Evanno et al., 2005), and by choosing the smallest K after the log probability of data [ln Pr(X|K)] values reached a plateau (Pritchard et al., 2010), using Structure Harvester (Earl and vonHoldt, 2012). C. Bayesian clustering analysis when K = 2 and K = 3. 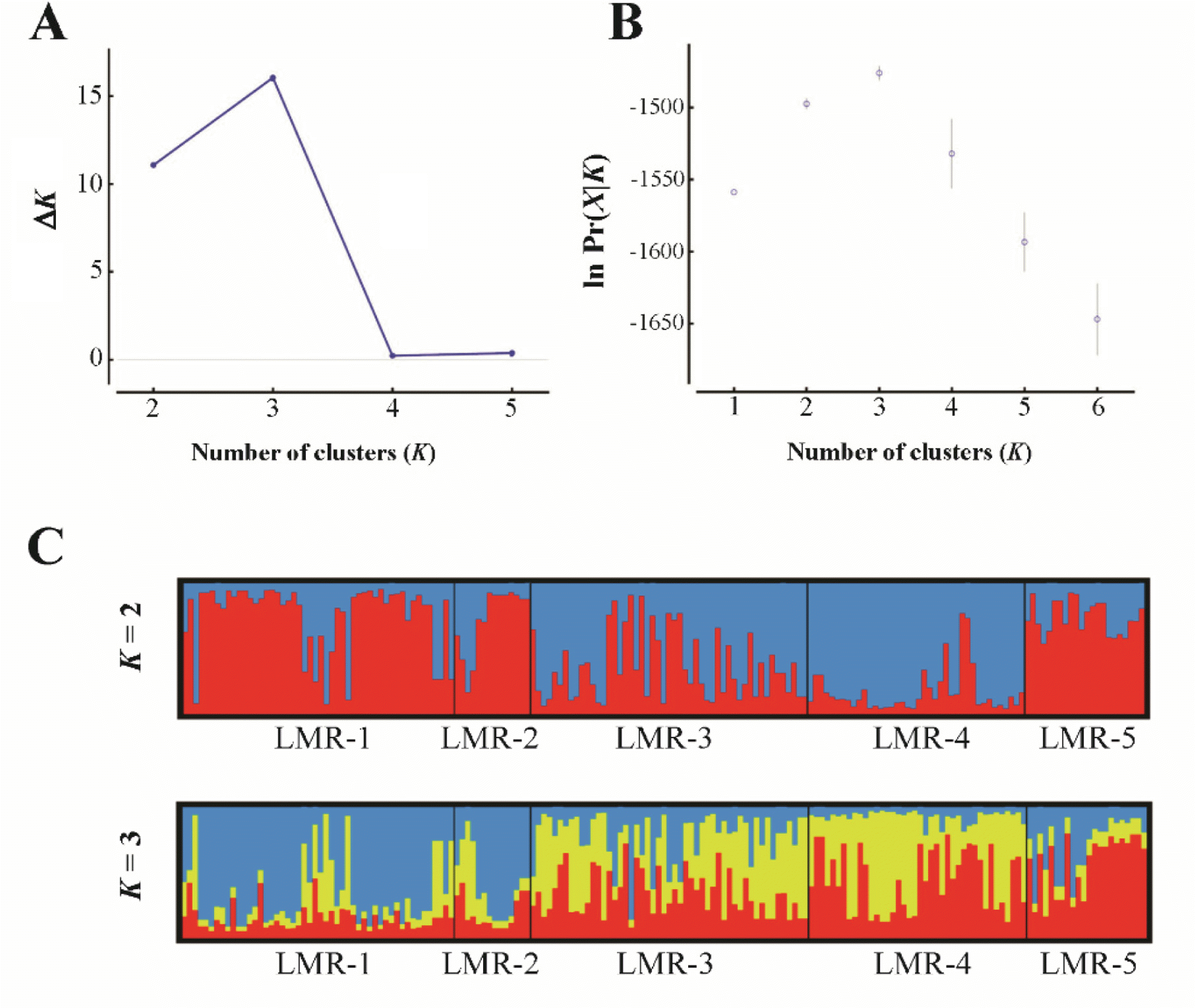
Table 1.Summary of within-population genetic diversity measures, mean fixation (FIS) and genetic divergence (GST or FST) estimates found in Liparis krameri, in two congeners (L. kumokiri and L. makinoana) in South Korea, and groups of species having similar-life history traits and species from areas recognized as harboring a glacial refugium (the BDDG).
n, the number of individuals; NP, number of populations; %P, percentage of polymorphic loci; A, mean number of alleles per locus; AR, mean allelic richness; Ho, observed heterozygosity; He, H–W expected heterozygosity or genetic diversity; SE, standard error in parentheses; N, number of studies (entries). Note that in the text a “P” subscript denotes population-level values (e.g., %PP, HeP), whereas an “S” subscript denotes species-level values (e.g., %PS, HeS). a Significant (at p = 0.000) deviation from the null hypothesis of FIS = 0 based on permutation (999 replicates). b Weir and Cockerham (1984) estimate of FIS over populations. c Significant (at p = 0.001) Weir and Cockerham (1984) estimate of FIS over populations. Literature CitedArditti, J and Ghani, AKA. 2000. Numerical and physical properties of orchid seeds and their biological implications. New Phytologist 145: 367-421.
Borzée, A. Santos, JL. Sánchez-Ramírez, S. Bae, Y. Heo, K. Jang, Y and Jowers, MJ. 2017. Phylogeographic and population insights of the Asian common toad (Bufo gargarizans) in Korea and China: population isolation and expansions as response to the ice ages. PeerJ 5: e4044.
Burczyk, J. Adams, WT. Birkes, DS and Chybicki, IJ. 2006. Using genetic markers to directly estimate gene flow and reproductive success parameters in plants on the basis of naturally regenerated seedlings. Genetics 173: 363-372.
Chen, X. Ormerod, P and Wood, JJ. 2009. Liparis Richard. In Flora of China, Vol. 25 (Orchidaceae). Wu, ZY. Raven, PH. Hong, DY (eds.), Science Press, Beijing and Missouri Botanical Garden Press, St. Louis, MO. 211-228.
Chung, MY. Nason, JD and Chung, MG. 2005. Patterns of hybridization and population genetic structure in the terrestrial orchids Liparis kumokiri and Liparis makinoana (Orchidaceae) in sympatric populations. Molecular Ecology 14: 4389-4402.
Chung, MY. López-Pujol, J and Chung, MG. 2017. The role of the Baekdudaegan (Korean Peninsula) as a major glacial refugium for plant species: a priority for conservation. Biological Conservation 206: 236-248.
Chung, MY. López-Pujol, J. Son, S. Suh, GU. Yukawa, T and Chung, MG. 2018b. Patterns of genetic diversity in rare and common orchids focusing on the Korean Peninsula: implications for conservation. The Botanical Review 84: 1-25.
Chung, MY. Moon, M-O. López-Pujol, J. Maki, M. Yamashiro, T. Yukawa, T. Sugiura, N. Lee, Y-I and Chung, MG. 2013. Was Jeju Island a glacial refugium for East Asian warm-temperate plants? Insights from the homosporous fern Selliguea hastata (Polypodiaceae). American Journal of Botany 100: 2240-2249.
Chung, MY. Park, C-W. Myers, ER and Chung, MG. 2007. Contrasting levels of genetic diversity between the common, self-compatible Liparis kumokiri and rare, self-incompatible Liparis makinoana (Orchidaceae) in South Korea. Botanical Journal of the Linnean Society 153: 41-48.
Chung, MY. Son, S. Suh, GU. Herrando-Moraira, S. Lee, CH. López-Pujol, J and Chung, MG. 2018a. The Korean Baekdudaegan Mountains: a glacial refugium and a biodiversity hotspot that needs to be conserved. Frontiers in Genetics 9: 489.
Cornuet, JM and Luikart, G. 1996. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144: 2001-2014.
Dolezal, J. Altman, J. Kopecky, M. Cerny, T. Janecek, S. Bartos, M. Petrik, P. Srutek, M. Leps, J and Song, J-S. 2012. Plant diversity changes during the postglacial in East Asia: insights from forest refugia on Halla volcano, Jeju Island. PLoS ONE 7: e33065.
Duminil, J. Hardy, OJ and Petit, RJ. 2009. Plant traits correlated with generation time directly affect inbreeding depression and mating system and indirectly genetic structure. BMC Evolutionary Biology 9: 177.
Earl, DA and vonHoldt, BM. 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4: 359-361.
El Mousadik, A and Petit, RJ. 1996. High level of genetic differentiation for allelic richness among populations of the argantree [Argania spinosa (L.) Skeels] endemic to Morocco. Theoretical and Applied Genetics 92: 832-839.
Evanno, G. Regnaut, S and Goudet, J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14: 2611-2620.
Gonzales, E. Hamrick, JL. Smouse, PE. Trapnell, DW and Peakall, R. 2010. The impact of landscape disturbance on spatial genetic structure in the Guanacaste tree, Enterolobium cyclocarpum (Fabaceae). Journal of Heredity 101: 133-143.
Goudet, J. 1995. FSTAT (version 1.2): a computer program to calculate F-statistics. Journal of Heredity 86: 485-486.
Grivet, D. Smouse, PE and Sork, VL. 2005. A novel approach to an old problem: tracking dispersed seeds. Molecular Ecology 14: 3585-3595.
Hamrick, JL. 2004. Response of forest trees to global environmental changes. Forest Ecology and Management 197: 323-335.
Hamrick, JL. Blanton, HM and Hamrick, KJ. 1989. The genetic structure of geographically marginal populations of ponderosa pine. American Journal of Botany 76: 1559-1568.
Hamrick, JL and Godt, MJW. 1989. Allozyme diversity in plant species. In Plant Population Genetics, Breeding and Genetic Resources. Brown, AHD. Clegg, MT. Kahler, AL. Weir, BS (eds.), Sinauer Associates, Sunderland, MA. 43-63.
Hamrick, JL and Godt, MJW. 1996. Effects of life history traits on genetic diversity in plant species. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences 351: 1291-1298.
Hardesty, BD. Hubbell, SP and Bermingham, E. 2006. Genetic evidence of frequent long-distance recruitment in a vertebrate-dispersed tree. Ecology Letters 9: 516-525.
Hardy, OJ and Vekemans, X. 2002. SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molecular Ecology Notes 2: 618-620.
Hurlbert, SH. 1971. The nonconcept of species diversity: a critique and alternative parameters. Ecology 52: 577-586.
Janes, JK. Miller, JM. Dupuis, JR. Malenfant, RM. Gorrell, JC. Cullingham, CI and Andrew, RL. 2017. The K = 2 conundrum. Molecular Ecology 26: 3594-3602.
Jordano, P. Garcia, C. Godoy, JA and García-Castaño, JL. 2007. Differential contribution of frugivores to complex seed dispersal patterns. Proceedings of the National Academy of Sciences of the United States of America 104: 3278-3282.
Loveless, MD and Hamrick, JL. 1984. Ecological determinants of genetic structure in plant populations. Annual Review of Ecology and Systematics 15: 65-95.
Luikart, G. Allendorf, FW. Cornuet, J-M and Sherwin, WB. 1998. Distortion of allele frequency distributions provides a test for recent population bottlenecks. Journal of Heredity 89: 238-247.
Mitton, JB. Linhart, YB. Sturgeon, KB and Hamrick, JL. 1979. Allozyme polymorphisms detected in mature needle tissue of ponderosa pine. Journal of Heredity 70: 86-89.
Nei, M. 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89: 583-590.
Nei, M. Maruyama, T and Chakraborty, R. 1975. The bottleneck effect and genetic variability in populations. Evolution 29: 1-10.
Nybom, H. 2004. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Molecular Ecology 13: 1143-1155.
Oh, GS. Chung, MY. Chung, SG and Chung, MG. 2001. Contrasting breeding systems: Liparis kumokiri and L. makinoana (Orchidaceae). Annales Botanici Fennici 38: 281-284.
Olson, MS. Hamrick, JL and Moore, RC. 2016. Breeding systems, mating systems, and gender determination in angiosperm trees. In Comparative and Evolutionary Genomics of Angiosperm Trees. Groover, A. Cronk, Q (eds.), Springer, Cham. 139-158.
Ottewell, KM. Bickerton, DC. Byrne, M and Lowe, AJ. 2016. Bridging the gap: a genetic assessment framework for population-level threatened plant conservation prioritization and decision-making. Diversity and Distributions 22: 174-188.
Peakall, R and Smouse, PE. 2012. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research: an update. Bioinformatics 28: 2537-2539.
Pritchard, JK. Stephens, M and Donnelly, P. 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945-959.
Pritchard, JK. Wen, X and Falush, D. 2010. Documentation for structure software: version 2.3. Retrieved Jul. 6, 2019, available from http://pritch.bsd.uchicago.edu/structure_software/release_versions/v2.3.4/structure_doc.pdf..
Ritland, K and Jain, S. 1981. A model for the estimation of outcrossing rate and gene frequencies using n independent loci. Heredity 47: 35-52.
Schnabel, A. Nason, JD and Hamrick, JL. 1998. Understanding the population genetic structure of Gleditsia triacanthos L.: seed dispersal and variation in female reproductive success. Molecular Ecology 7: 819-832.
Sezen, UU. Chazdon, RL and Holsinger, KE. 2009. Proximity is not a proxy for parentage in an animal-dispersed neotropical canopy palm. Proceedings of the Royal Society of London. Series B: Biological Sciences 276: 2037-2044.
Suetsugu, K. 2019. Rain-triggered self-pollination in Liparis kumokiri, an orchid that blooms during the rainy season. Ecology 100: e02683.
Takashima, M. Hasegawa, J and Yukawa, T. 2016. Oreorchis coreana (Orchidaceae), a new addition to the flora of Japan. Acta Phytotaxonomica et Geobotanica 67: 61-66.
Trapnell, DW and Hamrick, JL. 2004. Partitioning nuclear and chloroplast variation at multiple spatial scales in the neotropical epiphytic orchid, Laelia rubescens. Molecular Ecology 13: 2655-2666.
Troupin, D. Nathan, R and Vendramin, GG. 2006. Analysis of spatial genetic structure in an expanding Pinus halepensis population reveals development of fine-scale genetic clustering over time. Molecular Ecology 15: 3617-3630.
Vekemans, X and Hardy, OJ. 2004. New insights from fine-scale spatial genetic structure analyses in plant populations. Molecular Ecology 13: 921-935.
Weir, BS and Cockerham, CC. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38: 1358-1370.
Whitehead, MR. Lanfear, R. Mitchell, RJ and Karron, JD. 2018. Plant mating systems often vary widely among populations. Frontiers in Ecology and Evolution 6: 38.
Wright, S. 1965. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution 19: 395-420.
Yeh, FC. Yang, RC and Boyle, TBJ. 1999. POPGENE version 1.31: Microsoft Windows-based freeware for population genetic analysis. Quick Users’ Guide. University of Alberta, Edmonton.
Yukawa, T. Kawaguchi, D. Mukai, A and Komaki, Y. 2012. Discovery of Geodorum densiflorum (Orchidaceae) on the Ogasawara (Bonin) Islands: a case of ongoing colonization subsequent to long-distance dispersal. Bulletin of the National Museum of Nature and Science. Series B, Botany 38: 131-137.
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||